Identification and Characterization of Novel Maize Mirnas Involved in Different Genetic Background
- PMID: 26078720
- PMCID: PMC4466459
- DOI: 10.7150/ijbs.11619
Identification and Characterization of Novel Maize Mirnas Involved in Different Genetic Background
Abstract
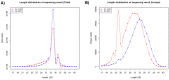
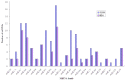
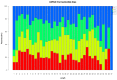
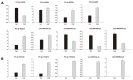
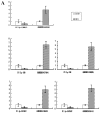
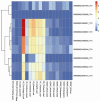
MicroRNAs (miRNAs) are a class of small, non-coding regulatory RNAs that regulate gene expression by guiding target mRNA cleavage or translational inhibition in plants and animals. At present there is relatively little information regarding the role of miRNAs in the response to drought stress in maize. In this study, two small RNA libraries were sequenced, and a total of 11,973,711 and 14,326,010 raw sequences were generated from growing leaves of drought-tolerant and drought-sensitive maize seedlings, respectively. Further analysis identified 192 mature miRNAs, which include 124 known maize (zma) miRNAs and 68 potential novel miRNA candidates. Additionally, 167 target genes (259 transcripts) of known and novel miRNAs were predicted to be differentially expressed between two maize inbred lines. Of these, three novel miRNAs were up-regulated and two were down-regulated under drought stress. The expression of these five miRNAs and nine target genes was confirmed using quantitative reverse transcription PCR. The expression of three of the miRNAs and their putative target genes exhibited an inverse correlation, and expression analysis suggested that all five may play important roles in maize leaves. Finally, GO annotations of the target genes indicated a potential role in photosynthesis, may therefore contribute to the drought stress response. This study describes the identification and characterization of novel miRNAs that are the differentially expressed in drought-tolerant and drought-sensitive inbred maize lines. This provides the foundation for further investigation into the mechanism of miRNA function in response to drought stress in maize.
Keywords: Drought stress; High-throughput sequencing; Maize; MicroRNA; Target genes; qRT-PCR.
Conflict of interest statement
Competing Interests: The authors have declared that no competing interest exists.
Figures

Similar articles
-
System analysis of microRNAs in the development and aluminium stress responses of the maize root system.Plant Biotechnol J. 2014 Oct;12(8):1108-21. doi: 10.1111/pbi.12218. Epub 2014 Jul 1. Plant Biotechnol J. 2014. PMID: 24985700
-
Genome-wide identification of microRNAs responding to early stages of phosphate deficiency in maize.Physiol Plant. 2016 Jun;157(2):161-74. doi: 10.1111/ppl.12409. Epub 2016 Jan 29. Physiol Plant. 2016. PMID: 26572939
-
Expression of zma-miR169 miRNAs and their target ZmNF-YA genes in response to abiotic stress in maize leaves.Gene. 2015 Jan 25;555(2):178-85. doi: 10.1016/j.gene.2014.11.001. Epub 2014 Nov 5. Gene. 2015. PMID: 25445264
-
Transcriptional regulatory networks in response to drought stress and rewatering in maize (Zea mays L.).Mol Genet Genomics. 2021 Nov;296(6):1203-1219. doi: 10.1007/s00438-021-01820-y. Epub 2021 Oct 3. Mol Genet Genomics. 2021. PMID: 34601650 Review.
-
Transcriptome expression profiles reveal response mechanisms to drought and drought-stress mitigation mechanisms by exogenous glycine betaine in maize.Biotechnol Lett. 2022 Mar;44(3):367-386. doi: 10.1007/s10529-022-03221-6. Epub 2022 Mar 16. Biotechnol Lett. 2022. PMID: 35294695 Review.
Cited by
-
ZmDST44 Gene Is a Positive Regulator in Plant Drought Stress Tolerance.Biology (Basel). 2024 Jul 23;13(8):552. doi: 10.3390/biology13080552. Biology (Basel). 2024. PMID: 39194490 Free PMC article.
-
Using Biotechnology-Led Approaches to Uplift Cereal and Food Legume Yields in Dryland Environments.Front Plant Sci. 2018 Aug 27;9:1249. doi: 10.3389/fpls.2018.01249. eCollection 2018. Front Plant Sci. 2018. PMID: 30210519 Free PMC article. Review.
-
Dehydration-responsive miRNAs in foxtail millet: genome-wide identification, characterization and expression profiling.Planta. 2016 Mar;243(3):749-66. doi: 10.1007/s00425-015-2437-7. Epub 2015 Dec 16. Planta. 2016. PMID: 26676987
-
Genome-wide identification of microRNAs responsive to Ectropis oblique feeding in tea plant (Camellia sinensis L.).Sci Rep. 2017 Oct 19;7(1):13634. doi: 10.1038/s41598-017-13692-7. Sci Rep. 2017. PMID: 29051614 Free PMC article.
-
Functional Roles of microRNAs in Agronomically Important Plants-Potential as Targets for Crop Improvement and Protection.Front Plant Sci. 2017 Mar 22;8:378. doi: 10.3389/fpls.2017.00378. eCollection 2017. Front Plant Sci. 2017. PMID: 28382044 Free PMC article. Review.
References
-
- Bartel DP. MicroRNAs: genomics, biogenesis, mechanism, and function. Cell. 2004;116:281–297. - PubMed
-
- Jones-Rhoades MW, Bartel DP, Bartel B. MicroRNAs and their regulatory roles in plants. Ann Rev Plant Biol. 2006;57:19–53. - PubMed
-
- Voinnet O. Origin, biogenesis, and activity of plant microRNAs. Cell. 2009;136:669–687. - PubMed
-
- Chuck G, Candela H, Hake S. Big impacts by small RNAs in plant development. Plant Biology. 2009;12:81–86. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

