The tethering of chromatin to the nuclear envelope supports nuclear mechanics
- PMID: 26074052
- PMCID: PMC4490570
- DOI: 10.1038/ncomms8159
The tethering of chromatin to the nuclear envelope supports nuclear mechanics
Abstract
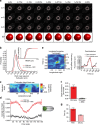
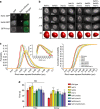
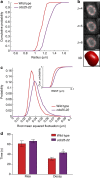
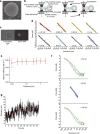
The nuclear lamina is thought to be the primary mechanical defence of the nucleus. However, the lamina is integrated within a network of lipids, proteins and chromatin; the interdependence of this network poses a challenge to defining the individual mechanical contributions of these components. Here, we isolate the role of chromatin in nuclear mechanics by using a system lacking lamins. Using novel imaging analyses, we observe that untethering chromatin from the inner nuclear membrane results in highly deformable nuclei in vivo, particularly in response to cytoskeletal forces. Using optical tweezers, we find that isolated nuclei lacking inner nuclear membrane tethers are less stiff than wild-type nuclei and exhibit increased chromatin flow, particularly in frequency ranges that recapitulate the kinetics of cytoskeletal dynamics. We suggest that modulating chromatin flow can define both transient and long-lived changes in nuclear shape that are biologically important and may be altered in disease.
Figures

Similar articles
-
Nuclear lamina at the crossroads of the cytoplasm and nucleus.J Struct Biol. 2012 Jan;177(1):24-31. doi: 10.1016/j.jsb.2011.11.007. Epub 2011 Nov 22. J Struct Biol. 2012. PMID: 22126840 Free PMC article. Review.
-
A lamin-independent pathway for nuclear envelope assembly.J Cell Biol. 1990 Dec;111(6 Pt 1):2247-59. doi: 10.1083/jcb.111.6.2247. J Cell Biol. 1990. PMID: 2277059 Free PMC article.
-
Nuclear reconstitution in vitro: stages of assembly around protein-free DNA.Cell. 1987 Jan 30;48(2):205-17. doi: 10.1016/0092-8674(87)90424-7. Cell. 1987. PMID: 3026635
-
Lamin activity is essential for nuclear envelope assembly in a Drosophila embryo cell-free extract.J Cell Biol. 1992 Oct;119(1):17-25. doi: 10.1083/jcb.119.1.17. J Cell Biol. 1992. PMID: 1527167 Free PMC article.
-
Review: nuclear lamins--structural proteins with fundamental functions.J Struct Biol. 2000 Apr;129(2-3):313-23. doi: 10.1006/jsbi.2000.4216. J Struct Biol. 2000. PMID: 10806082 Review.
Cited by
-
DNA content contributes to nuclear size control in Xenopus laevis.Mol Biol Cell. 2020 Nov 15;31(24):2703-2717. doi: 10.1091/mbc.E20-02-0113. Epub 2020 Sep 30. Mol Biol Cell. 2020. PMID: 32997613 Free PMC article.
-
Chromatin rigidity provides mechanical and genome protection.Mutat Res. 2020 May-Dec;821:111712. doi: 10.1016/j.mrfmmm.2020.111712. Epub 2020 Jun 17. Mutat Res. 2020. PMID: 32590202 Free PMC article. Review.
-
Defects in Emerin-Nucleoskeleton Binding Disrupt Nuclear Structure and Promote Breast Cancer Cell Motility and Metastasis.Mol Cancer Res. 2021 Jul;19(7):1196-1207. doi: 10.1158/1541-7786.MCR-20-0413. Epub 2021 Mar 26. Mol Cancer Res. 2021. PMID: 33771882 Free PMC article.
-
Migration through a small pore disrupts inactive chromatin organization in neutrophil-like cells.BMC Biol. 2018 Nov 26;16(1):142. doi: 10.1186/s12915-018-0608-2. BMC Biol. 2018. PMID: 30477489 Free PMC article.
-
PPAR-γ Agonist Pioglitazone Restored Mouse Liver mRNA Expression of Clock Genes and Inflammation-Related Genes Disrupted by Reversed Feeding.PPAR Res. 2022 May 26;2022:7537210. doi: 10.1155/2022/7537210. eCollection 2022. PPAR Res. 2022. PMID: 35663475 Free PMC article.
References
-
- Graumann K., Irons S. L., Runions J. & Evans D. E. Retention and mobility of the mammalian lamin B receptor in the plant nuclear envelope. Biol. Cell 99, 553–562 (2007). - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

