SlbHLH068 interacts with FER to regulate the iron-deficiency response in tomato
- PMID: 26070639
- PMCID: PMC4479748
- DOI: 10.1093/aob/mcv058
SlbHLH068 interacts with FER to regulate the iron-deficiency response in tomato
Abstract
Background and aims: Iron is an essential micronutrient for all organisms and its uptake, translocation, distribution and utilization are regulated in a complex manner in plants. FER, isolated from tomato (Solanum lycopersicum), was the first transcription factor involved in the iron homeostasis of higher plants to be identified. A FER defect in the T3238fer mutant drastically downregulates the expression of iron uptake genes, such as ferric-chelate reductase 1 (LeFRO1) and iron-regulated transporter 1 (LeIRT1); however, the molecular mechanism by which FER regulates genes downstream remains unknown. The aim of this work was therefore to identify the gene that interacts with FER to regulate the iron-deficiency response in tomato.
Methods: The homologue of the Arabidopsis Ib subgroup of the basic helix-loop-helix (bHLH) proteins, SlbHLH068, was identified by using the program BLASTP against the AtbHLH39 amino acid sequence in the tomato genome. The interaction between SlbHLH068 and FER was detected using yeast two-hybrid and bimolecular fluorescence complementation (BiFC) assays. In addition, virus-induced gene silencing (VIGS) was used to generate tomato plants in which SlbHLH068 expression was downregulated. The expression of genes was analysed using northern blot hybridization and multiple RT-PCR analysis. Seedlings of wild-type and mutant plants were grown under conditions of different nutrient deficiency.
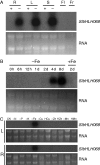
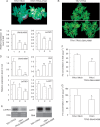
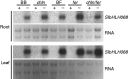
Key results: SlbHLH068 is highly upregulated in roots, leaves and stems in response to iron deficiency. An interaction between SlbHLH068 and FER was demonstrated using yeast two-hybrid and BiFC assays. The heterodimer formed by FER with SlbHLH068 directly bound to the promoter of LeFRO1 and activated the expression of its reporter gene in the yeast assay. The downregulation of SlbHLH068 expression by VIGS resulted in a reduction of LeFRO1 and LeIRT1 expression and iron accumulation in leaves and roots.
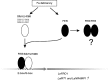
Conclusions: The results indicate that SlbHLH068, as a putative transcription factor, is involved in iron homeostasis in tomato via an interaction with FER.
Keywords: FER; SlbHLH068; Solanum lycopersicum; Tomato; ferric-chelate reductase 1; iron deficiency; iron homeostasis; iron-regulated transporter 1; transcription factor.
© The Author 2015. Published by Oxford University Press on behalf of the Annals of Botany Company. All rights reserved. For Permissions, please email: journals.permissions@oup.com.
Figures




Similar articles
-
Isolation and characterization of Fe(III)-chelate reductase gene LeFRO1 in tomato.Plant Mol Biol. 2004 Jan;54(1):125-36. doi: 10.1023/B:PLAN.0000028774.82782.16. Plant Mol Biol. 2004. PMID: 15159639
-
AtbHLH29 of Arabidopsis thaliana is a functional ortholog of tomato FER involved in controlling iron acquisition in strategy I plants.Cell Res. 2005 Aug;15(8):613-21. doi: 10.1038/sj.cr.7290331. Cell Res. 2005. PMID: 16117851
-
SlbHLH152, a bHLH transcription factor positively regulates iron homeostasis in tomato.Plant Sci. 2023 Oct;335:111821. doi: 10.1016/j.plantsci.2023.111821. Epub 2023 Aug 7. Plant Sci. 2023. PMID: 37558055
-
Nitric oxide accumulation is required for molecular and physiological responses to iron deficiency in tomato roots.Plant J. 2007 Dec;52(5):949-60. doi: 10.1111/j.1365-313X.2007.03283.x. Epub 2007 Sep 22. Plant J. 2007. PMID: 17892445
-
Twelve ways to confirm targets of activity-based probes in plants.Bioorg Med Chem. 2016 Aug 1;24(15):3304-11. doi: 10.1016/j.bmc.2016.05.036. Epub 2016 May 20. Bioorg Med Chem. 2016. PMID: 27298004 Review.
Cited by
-
The ethylene response factor AtERF4 negatively regulates the iron deficiency response in Arabidopsis thaliana.PLoS One. 2017 Oct 18;12(10):e0186580. doi: 10.1371/journal.pone.0186580. eCollection 2017. PLoS One. 2017. PMID: 29045490 Free PMC article.
-
Transcriptome Analysis Revealed the Molecular Response Mechanism of Non-heading Chinese Cabbage to Iron Deficiency Stress.Front Plant Sci. 2022 Mar 11;13:848424. doi: 10.3389/fpls.2022.848424. eCollection 2022. Front Plant Sci. 2022. PMID: 35371147 Free PMC article.
-
Downregulation of Zn-transporters along with Fe and redox imbalance causes growth and photosynthetic disturbance in Zn-deficient tomato.Sci Rep. 2021 Mar 16;11(1):6040. doi: 10.1038/s41598-021-85649-w. Sci Rep. 2021. PMID: 33727682 Free PMC article.
-
'Candidatus Phytoplasma solani' interferes with the distribution and uptake of iron in tomato.BMC Genomics. 2019 Sep 10;20(1):703. doi: 10.1186/s12864-019-6062-x. BMC Genomics. 2019. PMID: 31500568 Free PMC article.
-
Iron in the Symbiosis of Plants and Microorganisms.Plants (Basel). 2023 May 11;12(10):1958. doi: 10.3390/plants12101958. Plants (Basel). 2023. PMID: 37653875 Free PMC article. Review.
References
-
- Becker R, Grün M, Scholz G. 1992. Nicotianamine and the distribution of iron into the apoplasm and symplasm of tomato (Lycopersicon esculentum Mill.): I. Determination of the apoplasmic and symplasmic iron pools in roots and leaves of the cultivar Bonner Beste and its nicotianamine-less mutant chloronerva. Planta 187: 48–52. - PubMed
-
- Bereczky Z, Wang HY, Schubert V, Ganal M, Bauer P. 2003. Differential regulation of nramp and irt metal transporter genes in wild-type and iron uptake mutants of tomato. The Journal of Biological Chemistry 278: 24697–24704. - PubMed
-
- Chen JC, Jiang CZ, Gookin TE, Hunter DA, Clark DG, Reid MS. 2004. Chalcone synthase as a reporter in virus-induced gene silencing studies of flower senescence. Plant Molecular Biology 55: 521–530. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

