Comprehensive, Integrative Genomic Analysis of Diffuse Lower-Grade Gliomas
- PMID: 26061751
- PMCID: PMC4530011
- DOI: 10.1056/NEJMoa1402121
Comprehensive, Integrative Genomic Analysis of Diffuse Lower-Grade Gliomas
Abstract
Background: Diffuse low-grade and intermediate-grade gliomas (which together make up the lower-grade gliomas, World Health Organization grades II and III) have highly variable clinical behavior that is not adequately predicted on the basis of histologic class. Some are indolent; others quickly progress to glioblastoma. The uncertainty is compounded by interobserver variability in histologic diagnosis. Mutations in IDH, TP53, and ATRX and codeletion of chromosome arms 1p and 19q (1p/19q codeletion) have been implicated as clinically relevant markers of lower-grade gliomas.
Methods: We performed genomewide analyses of 293 lower-grade gliomas from adults, incorporating exome sequence, DNA copy number, DNA methylation, messenger RNA expression, microRNA expression, and targeted protein expression. These data were integrated and tested for correlation with clinical outcomes.
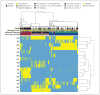
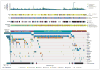
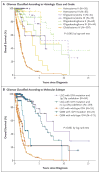
Results: Unsupervised clustering of mutations and data from RNA, DNA-copy-number, and DNA-methylation platforms uncovered concordant classification of three robust, nonoverlapping, prognostically significant subtypes of lower-grade glioma that were captured more accurately by IDH, 1p/19q, and TP53 status than by histologic class. Patients who had lower-grade gliomas with an IDH mutation and 1p/19q codeletion had the most favorable clinical outcomes. Their gliomas harbored mutations in CIC, FUBP1, NOTCH1, and the TERT promoter. Nearly all lower-grade gliomas with IDH mutations and no 1p/19q codeletion had mutations in TP53 (94%) and ATRX inactivation (86%). The large majority of lower-grade gliomas without an IDH mutation had genomic aberrations and clinical behavior strikingly similar to those found in primary glioblastoma.
Conclusions: The integration of genomewide data from multiple platforms delineated three molecular classes of lower-grade gliomas that were more concordant with IDH, 1p/19q, and TP53 status than with histologic class. Lower-grade gliomas with an IDH mutation either had 1p/19q codeletion or carried a TP53 mutation. Most lower-grade gliomas without an IDH mutation were molecularly and clinically similar to glioblastoma. (Funded by the National Institutes of Health.).
Figures



Comment in
-
Multiple Molecular Data Sets and the Classification of Adult Diffuse Gliomas.N Engl J Med. 2015 Jun 25;372(26):2555-7. doi: 10.1056/NEJMe1506813. Epub 2015 Jun 10. N Engl J Med. 2015. PMID: 26061754 No abstract available.
-
CNS cancer: molecular classification of glioma.Nat Rev Clin Oncol. 2015 Sep;12(9):502. doi: 10.1038/nrclinonc.2015.111. Epub 2015 Jul 14. Nat Rev Clin Oncol. 2015. PMID: 26169923 No abstract available.
Similar articles
-
TERT promoter mutation confers favorable prognosis regardless of 1p/19q status in adult diffuse gliomas with IDH1/2 mutations.Acta Neuropathol Commun. 2020 Nov 23;8(1):201. doi: 10.1186/s40478-020-01078-2. Acta Neuropathol Commun. 2020. PMID: 33228806 Free PMC article.
-
Glioma Groups Based on 1p/19q, IDH, and TERT Promoter Mutations in Tumors.N Engl J Med. 2015 Jun 25;372(26):2499-508. doi: 10.1056/NEJMoa1407279. Epub 2015 Jun 10. N Engl J Med. 2015. PMID: 26061753 Free PMC article.
-
RNA editing-based classification of diffuse gliomas: predicting isocitrate dehydrogenase mutation and chromosome 1p/19q codeletion.BMC Bioinformatics. 2019 Dec 24;20(Suppl 19):659. doi: 10.1186/s12859-019-3236-0. BMC Bioinformatics. 2019. PMID: 31870275 Free PMC article.
-
Biomarker-driven diagnosis of diffuse gliomas.Mol Aspects Med. 2015 Nov;45:87-96. doi: 10.1016/j.mam.2015.05.002. Epub 2015 May 21. Mol Aspects Med. 2015. PMID: 26004297 Review.
-
Integrated diagnostics of diffuse astrocytic and oligodendroglial tumors.Pathologe. 2019 Jun;40(Suppl 1):9-17. doi: 10.1007/s00292-019-0581-8. Pathologe. 2019. PMID: 31025086 Review. English.
Cited by
-
Glioblastoma, IDH-Wildtype Harboring a KIAA1549::BRAF Fusion: Report of a Case With Comprehensive Molecular Profiling.J Neuropathol Exp Neurol. 2022 Nov 16;81(12):1026-1028. doi: 10.1093/jnen/nlac076. J Neuropathol Exp Neurol. 2022. PMID: 36029520 Free PMC article. No abstract available.
-
Identification of Core Genes and Screening of Potential Targets in Glioblastoma Multiforme by Integrated Bioinformatic Analysis.Front Oncol. 2021 Feb 24;10:615976. doi: 10.3389/fonc.2020.615976. eCollection 2020. Front Oncol. 2021. PMID: 33718116 Free PMC article.
-
Systematic and Multi-Omics Prognostic Analysis of Lysine Acetylation Regulators in Glioma.Front Mol Biosci. 2021 Feb 26;8:587516. doi: 10.3389/fmolb.2021.587516. eCollection 2021. Front Mol Biosci. 2021. PMID: 33718432 Free PMC article.
-
Glioma progression is mediated by an addiction to aberrant IGFBP2 expression and can be blocked using anti-IGFBP2 strategies.J Pathol. 2016 Jul;239(3):355-64. doi: 10.1002/path.4734. Epub 2016 Jun 10. J Pathol. 2016. PMID: 27125842 Free PMC article.
-
Assessment of bevacizumab resistance increased by expression of BCAT1 in IDH1 wild-type glioblastoma: application of DSC perfusion MR imaging.Oncotarget. 2016 Oct 25;7(43):69606-69615. doi: 10.18632/oncotarget.11901. Oncotarget. 2016. PMID: 27626306 Free PMC article.
References
-
- Macdonald DR, Gaspar LE, Cairn-cross JG. Successful chemotherapy for newly diagnosed aggressive oligodendroglioma. Ann Neurol. 1990;27:573–4. - PubMed
-
- van den Bent MJ, Brandes AA, Taphoorn MJ, et al. Adjuvant procarbazine, lomustine, and vincristine chemotherapy in newly diagnosed anaplastic oligodendroglioma: long-term follow-up of EORTC brain tumor group study 26951. J Clin Oncol. 2013;31:344–50. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
- U24CA143840/CA/NCI NIH HHS/United States
- U24CA126561/CA/NCI NIH HHS/United States
- P30 CA016672/CA/NCI NIH HHS/United States
- U24 CA143882/CA/NCI NIH HHS/United States
- U54 HG003067/HG/NHGRI NIH HHS/United States
- U54HG003273/HG/NHGRI NIH HHS/United States
- U24 CA143835/CA/NCI NIH HHS/United States
- U24CA143731/CA/NCI NIH HHS/United States
- U24CA126551/CA/NCI NIH HHS/United States
- U24CA126543/CA/NCI NIH HHS/United States
- U24CA126554/CA/NCI NIH HHS/United States
- U24 CA143866/CA/NCI NIH HHS/United States
- U24CA143858/CA/NCI NIH HHS/United States
- U24CA126544/CA/NCI NIH HHS/United States
- P30CA016672/CA/NCI NIH HHS/United States
- U24CA143882/CA/NCI NIH HHS/United States
- U24 CA126551/CA/NCI NIH HHS/United States
- U24CA144025/CA/NCI NIH HHS/United States
- U54HG003067/HG/NHGRI NIH HHS/United States
- U24 CA143845/CA/NCI NIH HHS/United States
- U24 CA143799/CA/NCI NIH HHS/United States
- U01 CA168394/CA/NCI NIH HHS/United States
- U54 HG003273/HG/NHGRI NIH HHS/United States
- P30 CA008748/CA/NCI NIH HHS/United States
- U24 CA144025/CA/NCI NIH HHS/United States
- U24 CA194362/CA/NCI NIH HHS/United States
- U24CA143883/CA/NCI NIH HHS/United States
- U54HG003079/HG/NHGRI NIH HHS/United States
- U24 CA126554/CA/NCI NIH HHS/United States
- U24 CA180951/CA/NCI NIH HHS/United States
- U24 CA143840/CA/NCI NIH HHS/United States
- U24 CA143843/CA/NCI NIH HHS/United States
- U24CA143867/CA/NCI NIH HHS/United States
- U24CA143835/CA/NCI NIH HHS/United States
- U24 CA126561/CA/NCI NIH HHS/United States
- U24 CA143858/CA/NCI NIH HHS/United States
- U24CA143843/CA/NCI NIH HHS/United States
- R01 NS081125/NS/NINDS NIH HHS/United States
- U24 CA143848/CA/NCI NIH HHS/United States
- U24 CA126543/CA/NCI NIH HHS/United States
- U24CA143848/CA/NCI NIH HHS/United States
- U24CA126563/CA/NCI NIH HHS/United States
- U24CA126546/CA/NCI NIH HHS/United States
- U54 HG003079/HG/NHGRI NIH HHS/United States
- U24CA143799/CA/NCI NIH HHS/United States
- U54 HG007990/HG/NHGRI NIH HHS/United States
- U24 CA143883/CA/NCI NIH HHS/United States
- R01 CA163722/CA/NCI NIH HHS/United States
- U24CA143866/CA/NCI NIH HHS/United States
- U24CA143845/CA/NCI NIH HHS/United States
- U24 CA143867/CA/NCI NIH HHS/United States
- U24 CA126546/CA/NCI NIH HHS/United States
- U24 CA126563/CA/NCI NIH HHS/United States
- U24 CA126544/CA/NCI NIH HHS/United States
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials
Miscellaneous
