E. coli RNA Polymerase Determinants of Open Complex Lifetime and Structure
- PMID: 26055538
- PMCID: PMC4520765
- DOI: 10.1016/j.jmb.2015.05.024
E. coli RNA Polymerase Determinants of Open Complex Lifetime and Structure
Abstract
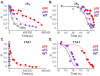
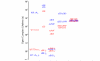
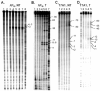
In transcription initiation by Escherichia coli RNA polymerase (RNAP), initial binding to promoter DNA triggers large conformational changes, bending downstream duplex DNA into the RNAP cleft and opening 13bp to form a short-lived open intermediate (I2). Subsequent conformational changes increase lifetimes of λPR and T7A1 open complexes (OCs) by >10(5)-fold and >10(2)-fold, respectively. OC lifetime is a target for regulation. To characterize late conformational changes, we determine effects on OC dissociation kinetics of deletions in RNAP mobile elements σ(70) region 1.1 (σ1.1), β' jaw and β' sequence insertion 3 (SI3). In very stable OC formed by the wild type WT RNAP with λPR (RPO) and by Δσ1.1 RNAP with λPR or T7A1, we conclude that downstream duplex DNA is bound to the jaw in an assembly with SI3, and bases -4 to +2 of the nontemplate strand discriminator region are stably bound in a positively charged track in the cleft. We deduce that polyanionic σ1.1 destabilizes OC by competing for binding sites in the cleft and on the jaw with the polyanionic discriminator strand and downstream duplex, respectively. Examples of σ1.1-destabilized OC are the final T7A1 OC and the λPR I3 intermediate OC. Deleting σ1.1 and either β' jaw or SI3 equalizes OC lifetimes for λPR and T7A1. DNA closing rates are similar for both promoters and all RNAP variants. We conclude that late conformational changes that stabilize OC, like early ones that bend the duplex into the cleft, are primary targets of regulation, while the intrinsic DNA opening/closing step is not.
Keywords: kinetics; mechanism; regulation; transcription initiation; σ(70) region 1.1.
Copyright © 2015 Elsevier Ltd. All rights reserved.
Figures

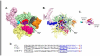

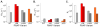
Similar articles
-
Clamp Interactions with +3/+6 Duplex and Upstream-to-Downstream Allosteric Effects in Late Steps of Forming a Stable RNA Polymerase-Promoter Open Complex.J Mol Biol. 2023 Mar 15;435(6):167990. doi: 10.1016/j.jmb.2023.167990. Epub 2023 Feb 1. J Mol Biol. 2023. PMID: 36736885
-
Mechanism of transcription initiation and promoter escape by E. coli RNA polymerase.Proc Natl Acad Sci U S A. 2017 Apr 11;114(15):E3032-E3040. doi: 10.1073/pnas.1618675114. Epub 2017 Mar 27. Proc Natl Acad Sci U S A. 2017. PMID: 28348246 Free PMC article.
-
Fluorescence-Detected Conformational Changes in Duplex DNA in Open Complex Formation by Escherichia coli RNA Polymerase: Upstream Wrapping and Downstream Bending Precede Clamp Opening and Insertion of the Downstream Duplex.Biochemistry. 2020 Apr 28;59(16):1565-1581. doi: 10.1021/acs.biochem.0c00098. Epub 2020 Apr 7. Biochemistry. 2020. PMID: 32216369 Free PMC article.
-
Initial events in bacterial transcription initiation.Biomolecules. 2015 May 27;5(2):1035-62. doi: 10.3390/biom5021035. Biomolecules. 2015. PMID: 26023916 Free PMC article. Review.
-
How sigma docks to RNA polymerase and what sigma does.Curr Opin Microbiol. 2001 Apr;4(2):126-31. doi: 10.1016/s1369-5274(00)00177-6. Curr Opin Microbiol. 2001. PMID: 11282466 Review.
Cited by
-
Structural origins of Escherichia coli RNA polymerase open promoter complex stability.Proc Natl Acad Sci U S A. 2021 Oct 5;118(40):e2112877118. doi: 10.1073/pnas.2112877118. Proc Natl Acad Sci U S A. 2021. PMID: 34599106 Free PMC article.
-
Macromolecular crowding has opposite effects on two critical sub-steps of transcription initiation.FEBS Lett. 2024 May;598(9):1022-1033. doi: 10.1002/1873-3468.14851. Epub 2024 Mar 13. FEBS Lett. 2024. PMID: 38479985 Free PMC article.
-
Diverse and unified mechanisms of transcription initiation in bacteria.Nat Rev Microbiol. 2021 Feb;19(2):95-109. doi: 10.1038/s41579-020-00450-2. Epub 2020 Oct 29. Nat Rev Microbiol. 2021. PMID: 33122819 Free PMC article. Review.
-
Delayed inhibition mechanism for secondary channel factor regulation of ribosomal RNA transcription.Elife. 2019 Feb 5;8:e40576. doi: 10.7554/eLife.40576. Elife. 2019. PMID: 30720429 Free PMC article.
-
Production and characterization of a highly pure RNA polymerase holoenzyme from Mycobacterium tuberculosis.Protein Expr Purif. 2017 Jun;134:1-10. doi: 10.1016/j.pep.2017.03.013. Epub 2017 Mar 18. Protein Expr Purif. 2017. PMID: 28323168 Free PMC article.
References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

