Members of the PpaA/AerR Antirepressor Family Bind Cobalamin
- PMID: 26055116
- PMCID: PMC4507344
- DOI: 10.1128/JB.00374-15
Members of the PpaA/AerR Antirepressor Family Bind Cobalamin
Abstract
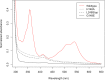
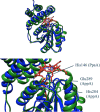
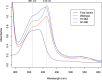
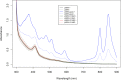
PpaA from Rhodobacter sphaeroides is a member of a family of proteins that are thought to function as antirepressors of PpsR, a widely disseminated repressor of photosystem genes in purple photosynthetic bacteria. PpaA family members exhibit sequence similarity to a previously defined SCHIC (sensor containing heme instead of cobalamin) domain; however, the tetrapyrrole-binding specificity of PpaA family members has been unclear, as R. sphaeroides PpaA has been reported to bind heme while the Rhodobacter capsulatus homolog has been reported to bind cobalamin. In this study, we reinvestigated tetrapyrrole binding of PpaA from R. sphaeroides and show that it is not a heme-binding protein but is instead a cobalamin-binding protein. We also use bacterial two-hybrid analysis to show that PpaA is able to interact with PpsR and activate the expression of photosynthesis genes in vivo. Mutations in PpaA that cause loss of cobalamin binding also disrupt PpaA antirepressor activity in vivo. We also tested a number of PpaA homologs from other purple bacterial species and found that cobalamin binding is a conserved feature among members of this family of proteins.
Importance: Cobalamin (vitamin B12) has only recently been recognized as a cofactor that affects gene expression by interacting in a light-dependent manner with transcription factors. A group of related antirepressors known as the AppA/PpaA/AerR family are known to control the expression of photosynthesis genes in part by interacting with either heme or cobalamin. The specificity of which tetrapyrroles that members of this family interact with has, however, remained cloudy. In this study, we address the tetrapyrrole-binding specificity of the PpaA/AerR subgroup and establish that it preferentially binds cobalamin over heme.
Copyright © 2015, American Society for Microbiology. All Rights Reserved.
Figures



Similar articles
-
The PpaA/AerR regulators of photosynthesis gene expression from anoxygenic phototrophic proteobacteria contain heme-binding SCHIC domains.J Bacteriol. 2010 Oct;192(19):5253-6. doi: 10.1128/JB.00736-10. Epub 2010 Jul 30. J Bacteriol. 2010. PMID: 20675482 Free PMC article.
-
Redox and light control the heme-sensing activity of AppA.mBio. 2013 Aug 27;4(5):e00563-13. doi: 10.1128/mBio.00563-13. mBio. 2013. PMID: 23982072 Free PMC article.
-
Identification and in vivo characterization of PpaA, a regulator of photosystem formation in Rhodobacter sphaeroides.Microbiology (Reading). 2003 Feb;149(Pt 2):377-388. doi: 10.1099/mic.0.25972-0. Microbiology (Reading). 2003. PMID: 12624200
-
PpsR: a multifaceted regulator of photosynthesis gene expression in purple bacteria.Mol Microbiol. 2005 Jul;57(1):17-26. doi: 10.1111/j.1365-2958.2005.04655.x. Mol Microbiol. 2005. PMID: 15948946 Review.
-
The tetrapyrrole biosynthetic pathway and its regulation in Rhodobacter capsulatus.Adv Exp Med Biol. 2010;675:229-50. doi: 10.1007/978-1-4419-1528-3_13. Adv Exp Med Biol. 2010. PMID: 20532744 Free PMC article. Review.
Cited by
-
A New Facet of Vitamin B12: Gene Regulation by Cobalamin-Based Photoreceptors.Annu Rev Biochem. 2017 Jun 20;86:485-514. doi: 10.1146/annurev-biochem-061516-044500. Annu Rev Biochem. 2017. PMID: 28654327 Free PMC article. Review.
-
Role of the CarH photoreceptor protein environment in the modulation of cobalamin photochemistry.Biophys J. 2021 Sep 7;120(17):3688-3696. doi: 10.1016/j.bpj.2021.07.020. Epub 2021 Jul 24. Biophys J. 2021. PMID: 34310939 Free PMC article.
-
A singular PpaA/AerR-like protein in Rhodospirillum rubrum rules beyond the boundaries of photosynthesis in response to the intracellular redox state.mSystems. 2023 Dec 21;8(6):e0070223. doi: 10.1128/msystems.00702-23. Epub 2023 Dec 6. mSystems. 2023. PMID: 38054698 Free PMC article.
-
The blue light-dependent LOV-protein LdaP of Dinoroseobacter shibae acts as antirepressor of the PpsR repressor, regulating photosynthetic gene cluster expression.Front Microbiol. 2024 Feb 7;15:1351297. doi: 10.3389/fmicb.2024.1351297. eCollection 2024. Front Microbiol. 2024. PMID: 38404597 Free PMC article.
-
Transcriptome and Deletion Mutant Analyses Revealed that an RpoH Family Sigma Factor Is Essential for Photosystem Production in Roseateles depolymerans under Carbon Starvation.Microbes Environ. 2023;38(1):ME22072. doi: 10.1264/jsme2.ME22072. Microbes Environ. 2023. PMID: 36878600 Free PMC article.
References
-
- Bauer CE, Setterdahl A, Wu J, Robinson BR. 2008. Regulation of gene expression in response to oxygen tension, p 707–725. In Hunter CN, Daldal F, Thurnauer MC, Beatty JT (ed), Purple photosynthetic bacteria. Kluwer Academic Press, Dordrecht, The Netherlands.
-
- Penfold RJ, Pemberton JM. 1991. A gene from the photosynthetic gene cluster of Rhodobacter sphaeroides induces trans-suppression of bacteriochlorophyll and carotenoid levels in R. sphaeroides and R. capsulatus. Curr Microbiol 23:259–263. doi:10.1007/BF02092027. - DOI
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

