Different KChIPs compete for heteromultimeric assembly with pore-forming Kv4 subunits
- PMID: 26039167
- PMCID: PMC4457479
- DOI: 10.1016/j.bpj.2015.04.024
Different KChIPs compete for heteromultimeric assembly with pore-forming Kv4 subunits
Abstract
Auxiliary Kv channel-interacting proteins 1-4 (KChIPs1-4) coassemble with pore-forming Kv4 α-subunits to form channel complexes underlying somatodendritic subthreshold A-type current that regulates neuronal excitability. It has been hypothesized that different KChIPs can competitively bind to Kv4 α-subunit to form variable channel complexes that can exhibit distinct biophysical properties for modulation of neural function. In this study, we use single-molecule subunit counting by total internal reflection fluorescence microscopy in combinations with electrophysiology and biochemistry to investigate whether different isoforms of auxiliary KChIPs, KChIP4a, and KChIP4bl, can compete for binding of Kv4.3 to coassemble heteromultimeric channel complexes for modulation of channel function. To count the number of photobleaching steps solely from cell membrane, we take advantage of a membrane tethered k-ras-CAAX peptide that anchors cytosolic KChIP4 proteins to the surface for reduction of background noise. Single-molecule subunit counting reveals that the number of KChIP4 isoforms in Kv4.3-KChIP4 complexes can vary depending on the KChIP4 expression level. Increasing the amount of KChIP4bl gradually reduces bleaching steps of KChIP4a isoform proteins, and vice versa. Further analysis of channel gating kinetics from different Kv4-KChIP4 subunit compositions confirms that both KChIP4a and KChIP4bl can modulate the channel complex function upon coassembly. Taken together, our findings show that auxiliary KChIPs can heteroassemble with Kv4 in a competitive manner to form heteromultimeric Kv4-KChIP4 channel complexes that are biophysically distinct and regulated under physiological or pathological conditions.
Copyright © 2015 Biophysical Society. Published by Elsevier Inc. All rights reserved.
Figures

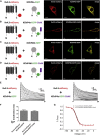
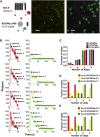

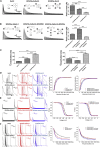
Similar articles
-
Multiple Kv channel-interacting proteins contain an N-terminal transmembrane domain that regulates Kv4 channel trafficking and gating.J Biol Chem. 2008 Dec 19;283(51):36046-59. doi: 10.1074/jbc.M806852200. Epub 2008 Oct 28. J Biol Chem. 2008. PMID: 18957440 Free PMC article.
-
Structural basis for modulation of Kv4 K+ channels by auxiliary KChIP subunits.Nat Neurosci. 2007 Jan;10(1):32-9. doi: 10.1038/nn1822. Epub 2006 Dec 24. Nat Neurosci. 2007. PMID: 17187064
-
The stoichiometry and biophysical properties of the Kv4 potassium channel complex with K+ channel-interacting protein (KChIP) subunits are variable, depending on the relative expression level.J Biol Chem. 2014 Jun 20;289(25):17597-609. doi: 10.1074/jbc.M114.563452. Epub 2014 May 8. J Biol Chem. 2014. PMID: 24811166 Free PMC article.
-
[Modulation of Kv4 channels by KChIPs clamping].Sheng Li Ke Xue Jin Zhan. 2009 Jan;40(1):9-13. Sheng Li Ke Xue Jin Zhan. 2009. PMID: 19408696 Review. Chinese.
-
Weighing the evidence for a ternary protein complex mediating A-type K+ currents in neurons.J Physiol. 2008 Dec 1;586(23):5609-23. doi: 10.1113/jphysiol.2008.161620. Epub 2008 Oct 9. J Physiol. 2008. PMID: 18845608 Free PMC article. Review.
Cited by
-
Phosphoinositide-interacting regulator of TRP (PIRT) has opposing effects on human and mouse TRPM8 ion channels.J Biol Chem. 2018 Jun 15;293(24):9423-9434. doi: 10.1074/jbc.RA118.003563. Epub 2018 May 3. J Biol Chem. 2018. PMID: 29724821 Free PMC article.
-
Characterisation of canine KCNIP4: A novel gene for cerebellar ataxia identified by whole-genome sequencing two affected Norwegian Buhund dogs.PLoS Genet. 2020 Jan 30;16(1):e1008527. doi: 10.1371/journal.pgen.1008527. eCollection 2020 Jan. PLoS Genet. 2020. PMID: 31999692 Free PMC article.
-
Number and Brightness Analysis: Visualization of Protein Oligomeric State in Living Cells.Adv Exp Med Biol. 2021;1310:31-58. doi: 10.1007/978-981-33-6064-8_2. Adv Exp Med Biol. 2021. PMID: 33834431
-
Kv4.2 and accessory dipeptidyl peptidase-like protein 10 (DPP10) subunit preferentially form a 4:2 (Kv4.2:DPP10) channel complex.J Biol Chem. 2015 Sep 11;290(37):22724-33. doi: 10.1074/jbc.M115.646794. Epub 2015 Jul 24. J Biol Chem. 2015. PMID: 26209633 Free PMC article.
-
Functional specification of CCK+ interneurons by alternative isoforms of Kv4.3 auxiliary subunits.Elife. 2020 Jun 3;9:e58515. doi: 10.7554/eLife.58515. Elife. 2020. PMID: 32490811 Free PMC article.
References
-
- An W.F., Bowlby M.R., Rhodes K.J. Modulation of A-type potassium channels by a family of calcium sensors. Nature. 2000;403:553–556. - PubMed
-
- Nadal M.S., Ozaita A., Rudy B. The CD26-related dipeptidyl aminopeptidase-like protein DPPX is a critical component of neuronal A-type K+ channels. Neuron. 2003;37:449–461. - PubMed
-
- Scannevin R.H., Wang K., Rhodes K.J. Two N-terminal domains of Kv4 K(+) channels regulate binding to and modulation by KChIP1. Neuron. 2004;41:587–598. - PubMed
-
- Cui Y.Y., Liang P., Wang K.W. Enhanced trafficking of tetrameric Kv4.3 channels by KChIP1 clamping. Neurochem. Res. 2008;33:2078–2084. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

