Systems-wide analysis of BCR signalosomes and downstream phosphorylation and ubiquitylation
- PMID: 26038114
- PMCID: PMC4501846
- DOI: 10.15252/msb.20145880
Systems-wide analysis of BCR signalosomes and downstream phosphorylation and ubiquitylation
Abstract
B-cell receptor (BCR) signaling is essential for the development and function of B cells; however, the spectrum of proteins involved in BCR signaling is not fully known. Here we used quantitative mass spectrometry-based proteomics to monitor the dynamics of BCR signaling complexes (signalosomes) and to investigate the dynamics of downstream phosphorylation and ubiquitylation signaling. We identify most of the previously known components of BCR signaling, as well as many proteins that have not yet been implicated in this system. BCR activation leads to rapid tyrosine phosphorylation and ubiquitylation of the receptor-proximal signaling components, many of which are co-regulated by both the modifications. We illustrate the power of multilayered proteomic analyses for discovering novel BCR signaling components by demonstrating that BCR-induced phosphorylation of RAB7A at S72 prevents its association with effector proteins and with endo-lysosomal compartments. In addition, we show that BCL10 is modified by LUBAC-mediated linear ubiquitylation, and demonstrate an important function of LUBAC in BCR-induced NF-κB signaling. Our results offer a global and integrated view of BCR signaling, and the provided datasets can serve as a valuable resource for further understanding BCR signaling networks.
Keywords: BCL10; BCR; RAB7A; phosphorylation; ubiquitylation.
© 2015 The Authors. Published under the terms of the CC BY 4.0 license.
Figures

Analysis of BCR-induced phosphorylation and ubiquitylation. A20 cells were isotopically labeled using the SILAC approach. Control “light” labeled cells were mock-treated, and “medium” and “heavy” labeled cells were stimulated with α-IgG F(ab′)2 for 5 and 15 min, respectively. Di-Gly-modified (ubiquitylated), tyrosine-phosphorylated peptides were enriched sequentially using di-Gly-lysine- and phosphotyrosine-specific antibodies. Phosphorylated peptides were separately enriched using TiO2-based chromatography. All samples were analyzed using high-resolution mass spectrometry.
Strategy for analyzing the dynamics of BCR signalosomes. “Medium” and “heavy” SILAC-labeled A20 cells were stimulated with biotinylated α-IgG F(ab′)2 for 5 and 15 min, respectively. Control cells (labeled with “light” SILAC) were mock-treated. Proteins associated with biotinylated α-IgG F(ab′)2-bound BCR signalosomes were affinity-enriched using streptavidin, separated by SDS–PAGE, and analyzed by LC-MS/MS.
Validation of BCR signaling activation. Stimulation of BCR signaling in A20 cells was confirmed using the indicated phosphorylation site-specific antibodies that recognize the activated forms of BTK, ERK1/2, and AKT kinases.

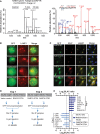
A The Venn diagram shows the overlap between the proteins that were enriched in BCR signalosomes at 5 and 15 min.
B A network view of proteins present in BCR signalosome. Proteins are color-coded based on their association with BCR signalosomes at 5 or 15 min after BCR cross-linking.
C Validation of the dynamic association of proteins with BCR signalosomes. A20 cells were stimulated with biotinylated α-IgG F(ab′)2 for the indicated time points, the signalosomes were isolated by streptavidin pull-downs, and enrichment of the indicated proteins was analyzed by immunoblotting.
D, E ANKRD13A associates with BCR signalosomes. A20 cells were transiently transfected with GFP-tagged ANKRD13A WT and the ANRKD13A ΔUIM mutant (D), or ANKRD13A WT and ANKRD13A UIM3/4 mutant (E). BCR signalosomes were isolated as described in (C), and the enrichment of ANKRD13A was probed using α-GFP antibody. The expression of GFP-tagged ANKRD13A WT, ANRKD13A ΔUIM, and ANKRD13A UIM3/4 mutants in the input material was verified.

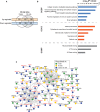
An overview of the BCR-regulated phosphoproteome. The table shows the number of the identified phosphorylation sites, and the number of sites that were up- (≥ SILAC ratio 2) or downregulated (≤ SILAC ratio 0.5) after 5 and 15 min of BCR stimulation.
Functional annotation of the BCR-upregulated phosphoproteome. The bar graph shows GO biological processes (GOBP) terms that were significantly enriched in BCR-upregulated phosphoproteome.
A diagram of the BCR signaling pathway. The proteins with upregulated phosphorylation sites in the pathway are colored, and the color gradient indicates the magnitude of phosphorylation increase. The modified amino acids (tyrosine or serine/threonine) are color-coded as circles, and the numbers of BCR-upregulated sites are indicated within the circles. Proteins with light yellow color were either not quantified or did not contain BCR-upregulated phosphorylation.
Identification of BCR-inducible RAB7A S72 phosphorylation. The MS spectrum (the left panel) shows the relative abundance of RAB7A S72 containing phosphopeptide at 5 and 15 min after the stimulation, and the MS/MS spectrum (the right panel) shows fragment ions of the identified peptide.
Subcellular localization of RAB7A phosphorylation mutants in B cells. Primary B cells were isolated from mouse spleen and infected with retroviruses expressing RAB7A WT or the indicated mutants fused with GFP. Cells were fixed with paraformaldehyde and stained with α-LAMP1, and epifluorescence images were acquired. Images show maximum projection of z-stacks of the cells.
Subcellular localization of RAB7A phosphorylation mutants in HeLa cells. The cells were co-transfected with FLAG-RILP and RAB7A or the indicated mutants fused with GFP. Cells were fixed, permeabilized, and stained with α-FLAG and α-LAMP1 antibodies to detect the exogenously expressed RILP and the endogenous LAMP1.
Strategy for identifying RAB7A interacting proteins. In experiment 1 (Exp 1), SILAC-labeled HeLa cells were transfected with GFP-RAB7A- or GFP-expressing plasmid vector to identify RAB7A interacting proteins. In a separate experiment (Exp 2), the interaction profile of GFP-RAB7A S72E was compared to GFP-RAB7A S72A.
Interaction profile of the RAB7A and the mutants. The bar graph shows the logarithmized SILAC ratios of the indicated RAB7A-binding proteins that were enriched (≥ 2-fold) with GFP-RAB7A compared to their levels in control immunoprecipitation (Exp 1), and their relative de-enrichment in GFP-RAB7A S72E compared to GFP-RAB7A S72A (Exp 2).
The overlap between BCR-regulated di-Gly-modified (ubiquitylation) sites. The Venn diagram shows the overlap of up- and downregulated sites after 5 or 15 min of BCR stimulation.
The functional annotation of the BCR-upregulated ubiquitylome. The bar graph indicates GO biological processes (GOBP), GO cellular compartments (GOCC), and GO molecular processes (GOMP) terms that were significantly enriched among the proteins containing BCR-upregulated di-Gly-modified sites.
Network analysis of the BCR-regulated ubiquitylome. The network shows proteins that contained BCR-upregulated di-Gly-modified sites at one or both of the time points. The proteins that also harbored BCR-induced phosphorylation are indicated in red type.
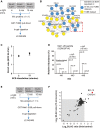
A Strategy for the identification of BCR-regulated ubiquitylated proteins. SILAC-labeled A20 cells were stimulated with α-IgG F(ab′)2 for the indicated times, and ubiquitylated proteins were affinity-enriched using tandem ubiquitin-binding entities (TUBE)-based pull-downs
B Interaction network of BCR-induced ubiquitylated proteins. The network shows interaction among the proteins that were significantly enriched in TUBE pull-downs from BCR-stimulated cells compared to proteins pulled down from mock-treated control cells. Blue circles indicate the proteins which were also identified as BCR-upregulated in our di-Gly dataset. The dotted box indicates members of the CBM complex.
C BCR stimulation increases the abundance of Met1-linked ubiquitin (Met1-UB). The plot shows the SILAC ratios of peptide corresponding to Met1-UB chains in the TUBE pull-downs from (A). The error bars represent mean ± SEM of the SILAC ratios.
D BCR stimulation increases the abundance of linear ubiquitin peptide. The MS spectrum shows the relative abundance of the peptide (GGMQIFVK) corresponding to Met1-UB in TUBE pull-downs from unstimulated cells, or after 5 or 15 min of BCR stimulation.
E, F Identification of BCR-regulated linear ubiquitylated proteins. Schematic presentation of the strategy used for SILAC-based Met1-SUB pull-downs (E). The scatter plot shows proteins identified in Met1-SUB pull-downs (F). The gray background indicates proteins that were identified in the pull-downs independent of BCR stimulation, and the red dots indicate proteins that were enriched after 5 and 15 min of BCR stimulation.

A, B Validation of BCR stimulation-dependent linear ubiquitylation of BCL10. BCL10 was immunoprecipitated from BCR-stimulated and unstimulated control A20 cells and immunoblotted with LUB9 antibody that binds to linear ubiquitin (A, the upper panel). From the same cell lysates, linear ubiquitylated proteins were isolated using Met1-SUB and probed with BCL10 (A, the lower panel). The latter approach was also used to confirm BCR-induced linear ubiquitylation of BCL10 in A20.2J cells (B). The blots at the bottom of the figure show the expression levels of BCL10 and actin in the input material used for BCL10 immunoprecipitation and Met1-SUB pull-downs.
C BCL10 ubiquitylation is sensitive to linear and K63 linkage-specific deubiquitylases. Linearly ubiquitylated proteins were isolated from BCR stimulated cells with Met1-SUB, treated with the indicated deubiquitylases, and subsequently immunostained with antibodies recognizing BCL10, or ubiquitin. The blots at the bottom of the figures show the expression levels of BCL10 and actin in the input material used for Met1-SUB pull-downs.

A HOIP interacts with BCL10. A20.2J HOIP−/− cells reconstituted with the full-length FLAG-HOIP were stimulated with α-IgG for the indicated times, HOIP was immunoprecipitated with α-FLAG antibody, and the immunoprecipitates were immunostained with BCL10 antibody. The amounts of immunoprecipitated HOIP, and equal expression of FLAG-HOIP and BCL10 in whole-cell lysates used for the IPs, are shown in the lower blots. The asterisk indicates unmodified BCL10.
B HOIP is required for BCL10 linear ubiquitylation. A20.2J, A20.2J HOIP−/−, and A20.2J HOIP−/− cells expressing the HOIP full-length or the indicated HOIP mutants were stimulated with α-IgG followed by Met1-SUB pull-down and immunoblotting as described in Fig7A.
C TRAF6 is required for BCL10 linear ubiquitylation. A20.2J wild-type and TRAF6−/− cells were stimulated with α-IgG for 10 min, and linear ubiquitylated proteins were pulled down with Met1-SUB and immunoblotted with BCL10 and ubiquitin antibodies. Equal expression of BCL10 and actin was verified in the input material.
D HOIP and TRAF6 function is important for BCR-induced IκB phosphorylation. A20.2J wild-type, HOIP−/−, TRAF6−/−, and HOIP−/− cells expressing HOIP ΔRBR were stimulated with α-IgG for the indicated time points, and lysates were immunostained with pIκB and IκB antibodies. Actin and vinculin staining serves as loading control.
E, F BCL10 linear ubiquitin fusion protein activates NF-κB. HEK293T cells were co-transfected with increasing amount (0.5, 1 or 2 μg) of BCL10, or BCL10-LinUBL73P-4X construct together with pNF-κB Luc and pRL-TK Renilla. NF-κB transcriptional activity was measured 24 h later using Dual-Glo® Luciferase Assay System (Promega). In (F), NF-κB transcriptional activity was measured in HEK293T cells co-transfected with 1 μg of the indicated plasmids. Error bars indicate mean ± SEM of 3 (E) or 2 (F) independent experiments. Statistical significance was determined by two-tailed Student's t-test.
Similar articles
-
The paracaspase MALT1 cleaves HOIL1 reducing linear ubiquitination by LUBAC to dampen lymphocyte NF-κB signalling.Nat Commun. 2015 Nov 3;6:8777. doi: 10.1038/ncomms9777. Nat Commun. 2015. PMID: 26525107 Free PMC article.
-
The novel adaptor protein Swiprosin-1 enhances BCR signals and contributes to BCR-induced apoptosis.Cell Death Differ. 2007 Nov;14(11):1936-47. doi: 10.1038/sj.cdd.4402206. Epub 2007 Aug 3. Cell Death Differ. 2007. PMID: 17673920
-
Processing of CD74 by the Intramembrane Protease SPPL2a Is Critical for B Cell Receptor Signaling in Transitional B Cells.J Immunol. 2015 Aug 15;195(4):1548-63. doi: 10.4049/jimmunol.1403171. Epub 2015 Jul 8. J Immunol. 2015. PMID: 26157172
-
Post-translational modifications regulate distinct functions of CARMA1 and BCL10.Trends Immunol. 2007 Jun;28(6):281-8. doi: 10.1016/j.it.2007.04.004. Epub 2007 Apr 30. Trends Immunol. 2007. PMID: 17468049 Review.
-
NF-kappaB signaling in lymphocytes: a new cast of characters.J Cell Sci. 2004 Jan 1;117(Pt 1):31-9. doi: 10.1242/jcs.00904. J Cell Sci. 2004. PMID: 14657271 Review.
Cited by
-
The immunobiology of ubiquitin-dependent B cell receptor functions.Mol Immunol. 2018 Sep;101:146-154. doi: 10.1016/j.molimm.2018.05.022. Epub 2018 Jun 26. Mol Immunol. 2018. PMID: 29940407 Free PMC article. Review.
-
Endophilin A2 regulates B-cell endocytosis and is required for germinal center and humoral responses.EMBO Rep. 2021 Sep 6;22(9):e51328. doi: 10.15252/embr.202051328. Epub 2021 Jul 29. EMBO Rep. 2021. PMID: 34323351 Free PMC article.
-
Illuminating Spatial and Temporal Organization of Protein Interaction Networks by Mass Spectrometry-Based Proteomics.Front Genet. 2015 Dec 1;6:344. doi: 10.3389/fgene.2015.00344. eCollection 2015. Front Genet. 2015. PMID: 26648978 Free PMC article. Review.
-
Serine 165 phosphorylation of SHARPIN regulates the activation of NF-κB.iScience. 2020 Dec 13;24(1):101939. doi: 10.1016/j.isci.2020.101939. eCollection 2021 Jan 22. iScience. 2020. PMID: 33392484 Free PMC article.
-
Molecular Determinants of Scaffold-induced Linear Ubiquitinylation of B Cell Lymphoma/Leukemia 10 (Bcl10) during T Cell Receptor and Oncogenic Caspase Recruitment Domain-containing Protein 11 (CARD11) Signaling.J Biol Chem. 2016 Dec 9;291(50):25921-25936. doi: 10.1074/jbc.M116.754028. Epub 2016 Oct 24. J Biol Chem. 2016. PMID: 27777308 Free PMC article.
References
-
- Bensimon A, Heck AJ, Aebersold R. Mass spectrometry-based proteomics and network biology. Annu Rev Biochem. 2012;81:379–405. - PubMed
-
- Brdicka T, Pavlistova D, Leo A, Bruyns E, Korinek V, Angelisova P, Scherer J, Shevchenko A, Hilgert I, Cerny J, Drbal K, Kuramitsu Y, Kornacker B, Horejsi V, Schraven B. Phosphoprotein associated with glycosphingolipid-enriched microdomains (PAG), a novel ubiquitously expressed transmembrane adaptor protein, binds the protein tyrosine kinase csk and is involved in regulation of T cell activation. J Exp Med. 2000;191:1591–1604. - PMC - PubMed
-
- Brems H, Chmara M, Sahbatou M, Denayer E, Taniguchi K, Kato R, Somers R, Messiaen L, De Schepper S, Fryns JP, Cools J, Marynen P, Thomas G, Yoshimura A, Legius E. Germline loss-of-function mutations in SPRED1 cause a neurofibromatosis 1-like phenotype. Nat Genet. 2007;39:1120–1126. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials

