Evolutionary comparison reveals that diverging CTCF sites are signatures of ancestral topological associating domains borders
- PMID: 26034287
- PMCID: PMC4475986
- DOI: 10.1073/pnas.1505463112
Evolutionary comparison reveals that diverging CTCF sites are signatures of ancestral topological associating domains borders
Abstract
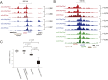
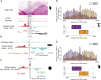
Increasing evidence in the last years indicates that the vast amount of regulatory information contained in mammalian genomes is organized in precise 3D chromatin structures. However, the impact of this spatial chromatin organization on gene expression and its degree of evolutionary conservation is still poorly understood. The Six homeobox genes are essential developmental regulators organized in gene clusters conserved during evolution. Here, we reveal that the Six clusters share a deeply evolutionarily conserved 3D chromatin organization that predates the Cambrian explosion. This chromatin architecture generates two largely independent regulatory landscapes (RLs) contained in two adjacent topological associating domains (TADs). By disrupting the conserved TAD border in one of the zebrafish Six clusters, we demonstrate that this border is critical for preventing competition between promoters and enhancers located in separated RLs, thereby generating different expression patterns in genes located in close genomic proximity. Moreover, evolutionary comparison of Six-associated TAD borders reveals the presence of CCCTC-binding factor (CTCF) sites with diverging orientations in all studied deuterostomes. Genome-wide examination of mammalian HiC data reveals that this conserved CTCF configuration is a general signature of TAD borders, underscoring that common organizational principles underlie TAD compartmentalization in deuterostome evolution.
Keywords: CTCF; Six cluster; TAD; evolution; regulatory landscapes.
Conflict of interest statement
The authors declare no conflict of interest.
Figures




Similar articles
-
5C analysis of the Epidermal Differentiation Complex locus reveals distinct chromatin interaction networks between gene-rich and gene-poor TADs in skin epithelial cells.PLoS Genet. 2017 Sep 1;13(9):e1006966. doi: 10.1371/journal.pgen.1006966. eCollection 2017 Sep. PLoS Genet. 2017. PMID: 28863138 Free PMC article.
-
Tissue-specific CTCF-cohesin-mediated chromatin architecture delimits enhancer interactions and function in vivo.Nat Cell Biol. 2017 Aug;19(8):952-961. doi: 10.1038/ncb3573. Epub 2017 Jul 24. Nat Cell Biol. 2017. PMID: 28737770 Free PMC article.
-
Sequential in cis mutagenesis in vivo reveals various functions for CTCF sites at the mouse HoxD cluster.Genes Dev. 2021 Nov 1;35(21-22):1490-1509. doi: 10.1101/gad.348934.121. Epub 2021 Oct 28. Genes Dev. 2021. PMID: 34711654 Free PMC article.
-
Topological Domains, Metagenes, and the Emergence of Pleiotropic Regulations at Hox Loci.Curr Top Dev Biol. 2016;116:299-314. doi: 10.1016/bs.ctdb.2015.11.022. Epub 2016 Jan 27. Curr Top Dev Biol. 2016. PMID: 26970625 Review.
-
Topologically associated domains: a successful scaffold for the evolution of gene regulation in animals.Wiley Interdiscip Rev Dev Biol. 2017 May;6(3). doi: 10.1002/wdev.265. Epub 2017 Mar 2. Wiley Interdiscip Rev Dev Biol. 2017. PMID: 28251841 Review.
Cited by
-
CTCF: making the right connections.Genes Dev. 2016 Apr 15;30(8):881-91. doi: 10.1101/gad.277863.116. Genes Dev. 2016. PMID: 27083996 Free PMC article. Review.
-
Escape From X-Chromosome Inactivation: An Evolutionary Perspective.Front Cell Dev Biol. 2019 Oct 22;7:241. doi: 10.3389/fcell.2019.00241. eCollection 2019. Front Cell Dev Biol. 2019. PMID: 31696116 Free PMC article. Review.
-
Uncovering direct and indirect molecular determinants of chromatin loops using a computational integrative approach.PLoS Comput Biol. 2017 May 23;13(5):e1005538. doi: 10.1371/journal.pcbi.1005538. eCollection 2017 May. PLoS Comput Biol. 2017. PMID: 28542178 Free PMC article.
-
Barcelona conference on epigenetics and cancer 2015: Coding and non-coding functions of the genome.Epigenetics. 2016;11(1):95-100. doi: 10.1080/15592294.2015.1131377. Epigenetics. 2016. PMID: 26996885 Free PMC article.
-
Molecular mechanism of directional CTCF recognition of a diverse range of genomic sites.Cell Res. 2017 Nov;27(11):1365-1377. doi: 10.1038/cr.2017.131. Epub 2017 Oct 27. Cell Res. 2017. PMID: 29076501 Free PMC article.
References
Publication types
MeSH terms
Substances
Associated data
- Actions
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials
Miscellaneous

