Insulin/IGF1-PI3K-dependent nucleolar localization of a glycolytic enzyme--phosphoglycerate mutase 2, is necessary for proper structure of nucleolus and RNA synthesis
- PMID: 26033454
- PMCID: PMC4627304
- DOI: 10.18632/oncotarget.4044
Insulin/IGF1-PI3K-dependent nucleolar localization of a glycolytic enzyme--phosphoglycerate mutase 2, is necessary for proper structure of nucleolus and RNA synthesis
Abstract
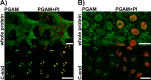
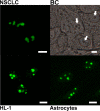
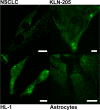
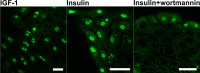
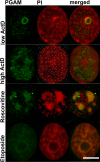
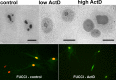
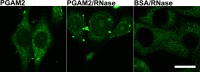
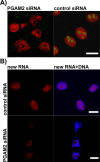
Phosphoglycerate mutase (PGAM), a conserved, glycolytic enzyme has been found in nucleoli of cancer cells. Here, we present evidence that accumulation of PGAM in the nucleolus is a universal phenomenon concerning not only neoplastically transformed but also non-malignant cells. Nucleolar localization of the enzyme is dependent on the presence of the PGAM2 (muscle) subunit and is regulated by insulin/IGF-1-PI3K signaling pathway as well as drugs influencing ribosomal biogenesis. We document that PGAM interacts with several 40S and 60S ribosomal proteins and that silencing of PGAM2 expression results in disturbance of nucleolar structure, inhibition of RNA synthesis and decrease of the mitotic index of squamous cell carcinoma cells. We conclude that presence of PGAM in the nucleolus is a prerequisite for synthesis and initial assembly of new pre-ribosome subunits.
Keywords: PGAM2; multifunctional enzyme; rRNA; ribosome assembly; squamous cell carcinoma.
Conflict of interest statement
The authors declare no conflict of interest.
Figures
Similar articles
-
High-Resolution Crystal Structure of Muscle Phosphoglycerate Mutase Provides Insight into Its Nuclear Import and Role.Int J Mol Sci. 2022 Oct 30;23(21):13198. doi: 10.3390/ijms232113198. Int J Mol Sci. 2022. PMID: 36361985 Free PMC article.
-
Oxidative stress activates SIRT2 to deacetylate and stimulate phosphoglycerate mutase.Cancer Res. 2014 Jul 1;74(13):3630-42. doi: 10.1158/0008-5472.CAN-13-3615. Epub 2014 May 1. Cancer Res. 2014. PMID: 24786789 Free PMC article.
-
Raptor, a positive regulatory subunit of mTOR complex 1, is a novel phosphoprotein of the rDNA transcription machinery in nucleoli and chromosomal nucleolus organizer regions (NORs).Cell Cycle. 2011 Sep 15;10(18):3140-52. doi: 10.4161/cc.10.18.17376. Epub 2011 Sep 15. Cell Cycle. 2011. PMID: 21900751
-
[Research progress in phosphoglycerate mutase].Zhongguo Xue Xi Chong Bing Fang Zhi Za Zhi. 2012 Jun;24(3):353-7. Zhongguo Xue Xi Chong Bing Fang Zhi Za Zhi. 2012. PMID: 23012968 Review. Chinese.
-
[Dynamics and mechanisms of the nucleolus reorganization during mitosis].Tsitologiia. 2007;49(5):355-69. Tsitologiia. 2007. PMID: 17654824 Review. Russian.
Cited by
-
Targeting signaling and apoptotic pathways involved in chemotherapeutic drug-resistance of hematopoietic cells.Oncotarget. 2017 Aug 24;8(44):76525-76557. doi: 10.18632/oncotarget.20408. eCollection 2017 Sep 29. Oncotarget. 2017. PMID: 29100331 Free PMC article.
-
Cell-to-cell lactate shuttle operates in heart and is important in age-related heart failure.Aging (Albany NY). 2020 Feb 8;12(4):3388-3406. doi: 10.18632/aging.102818. Epub 2020 Feb 8. Aging (Albany NY). 2020. PMID: 32035422 Free PMC article.
-
Nucleolar Proteins and Non-Coding RNAs: Roles in Renal Cancer.Int J Mol Sci. 2021 Dec 4;22(23):13126. doi: 10.3390/ijms222313126. Int J Mol Sci. 2021. PMID: 34884928 Free PMC article. Review.
-
High-Resolution Crystal Structure of Muscle Phosphoglycerate Mutase Provides Insight into Its Nuclear Import and Role.Int J Mol Sci. 2022 Oct 30;23(21):13198. doi: 10.3390/ijms232113198. Int J Mol Sci. 2022. PMID: 36361985 Free PMC article.
-
TMT-based quantitative proteomics analysis reveals the key proteins related with the differentiation process of goat intramuscular adipocytes.BMC Genomics. 2021 Jun 5;22(1):417. doi: 10.1186/s12864-021-07730-y. BMC Genomics. 2021. PMID: 34090334 Free PMC article.
References
-
- Zhang J, Yu L, Fu Q, Gao J, Xie Y, Chen J, Zhang P, Liu Q, Zhao S. Mouse phosphoglycerate mutase M and B isozymes: cDNA cloning, enzyme activity assay and mapping. Gene. 2001;264:273–9. - PubMed
-
- Kowalski W, Nocon D, Gamian A, Kolodziej J, Rakus D. Association of C-terminal region of phosphoglycerate mutase with glycolytic complex regulates energy production in cancer cells. J Cell Physiol. 2012;227:2613–2621. - PubMed
-
- Boisvert FM, van Koningsbruggen S, Navascués J, Lamond AI. The multifunctional nucleolus. Nat Rev Mol Cell Biol. 2007;8:574–585. - PubMed
-
- Takeda K, Komuro Y, Hayakawa T, Oguchi H, Ishida Y, Murakami S, Noguchi T, Kinoshita H, Sekine Y, Iemura S, Natsume T, Ichijo H. Mitochondrial phosphoglycerate mutase 5 uses alternate catalytic activity as a protein serine/threonine phosphatase to activate ASK1. Proc Natl Acad Sci U S A. 2009;106:12301–12305. - PMC - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

