Defining bacterial regulons using ChIP-seq
- PMID: 26032817
- PMCID: PMC4577457
- DOI: 10.1016/j.ymeth.2015.05.022
Defining bacterial regulons using ChIP-seq
Abstract
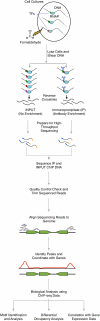
Chromatin immunoprecipitation followed by high-throughput sequencing (ChIP-seq) is a powerful method that identifies protein-DNA binding sites in vivo. Recent studies have illustrated the value of ChIP-seq in studying transcription factor binding in various bacterial species under a variety of growth conditions. These results show that in addition to identifying binding sites, correlation of ChIP-seq data with expression data can reveal important information about bacterial regulons and regulatory networks. In this chapter, we provide an overview of the current state of knowledge about ChIP-seq methodology in bacteria, from sample preparation to raw data analysis. We also describe visualization and various bioinformatic analyses of processed ChIP-seq data.
Keywords: Bacterial regulons; Bioinformatics analysis of genomic data; ChIP-seq; Genome-wide analysis; Systems biology; Transcription factor binding sites; Transcriptional regulation.
Copyright © 2015 Elsevier Inc. All rights reserved.
Figures

Similar articles
-
Technical considerations in using DNA microarrays to define regulons.Methods. 2009 Jan;47(1):63-72. doi: 10.1016/j.ymeth.2008.10.017. Epub 2008 Oct 26. Methods. 2009. PMID: 18955146 Free PMC article.
-
ChIP-seq for genome-scale analysis of bacterial DNA-binding proteins.Methods Mol Biol. 2015;1276:327-40. doi: 10.1007/978-1-4939-2392-2_20. Methods Mol Biol. 2015. PMID: 25665574
-
Direct ChIP-Seq significance analysis improves target prediction.BMC Genomics. 2015;16 Suppl 5(Suppl 5):S4. doi: 10.1186/1471-2164-16-S5-S4. Epub 2015 May 26. BMC Genomics. 2015. PMID: 26040656 Free PMC article.
-
Visualizing and characterizing in vivo DNA-binding events and direct target genes of plant transcription factors.Methods Mol Biol. 2011;754:293-305. doi: 10.1007/978-1-61779-154-3_17. Methods Mol Biol. 2011. PMID: 21720960 Review.
-
ChIP-Seq Data Analysis to Define Transcriptional Regulatory Networks.Adv Biochem Eng Biotechnol. 2017;160:1-14. doi: 10.1007/10_2016_43. Adv Biochem Eng Biotechnol. 2017. PMID: 28070596 Review.
Cited by
-
Exposing the small protein load of bacterial life.FEMS Microbiol Rev. 2023 Nov 1;47(6):fuad063. doi: 10.1093/femsre/fuad063. FEMS Microbiol Rev. 2023. PMID: 38012116 Free PMC article. Review.
-
Systematic mapping of protein-metabolite interactions in central metabolism of Escherichia coli.Mol Syst Biol. 2019 Aug;15(8):e9008. doi: 10.15252/msb.20199008. Mol Syst Biol. 2019. PMID: 31464375 Free PMC article.
-
Elucidating the functional roles of prokaryotic proteins using big data and artificial intelligence.FEMS Microbiol Rev. 2023 Jan 16;47(1):fuad003. doi: 10.1093/femsre/fuad003. FEMS Microbiol Rev. 2023. PMID: 36725215 Free PMC article. Review.
-
proChIPdb: a chromatin immunoprecipitation database for prokaryotic organisms.Nucleic Acids Res. 2022 Jan 7;50(D1):D1077-D1084. doi: 10.1093/nar/gkab1043. Nucleic Acids Res. 2022. PMID: 34791440 Free PMC article.
-
The primary σ factor in Escherichia coli can access the transcription elongation complex from solution in vivo.Elife. 2015 Sep 15;4:e10514. doi: 10.7554/eLife.10514. Elife. 2015. PMID: 26371553 Free PMC article.
References
-
- Browning DF, Busby SJ. The regulation of bacterial transcription initiation. Nat Rev Micro. 2004;2:57–65. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

