A chemical and computational approach to comprehensive glycation characterization on antibodies
- PMID: 26030340
- PMCID: PMC4622828
- DOI: 10.1080/19420862.2015.1046663
A chemical and computational approach to comprehensive glycation characterization on antibodies
Abstract
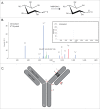
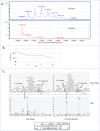
Non-enzymatic glycation is a challenging post-translational modification to characterize due to the structural heterogeneity it generates in proteins. Glycation has become increasingly recognized as an important product quality attribute to monitor, particularly for the biotechnology sector, which produces recombinant proteins under conditions that are amenable to protein glycation. The elucidation of sites of glycation can be problematic using conventional collision-induced dissociation (CID)-based mass spectrometry because of the predominance of neutral loss ions. A method to characterize glycation using an IgG1 monoclonal antibody (mAb) as a model is reported here. The sugars present on this mAb were derivatized using sodium borohydride chemistry to stabilize the linkage and identified using CID-based MS(2) mass spectrometry and spectral search engines. Quantification of specific glycation sites was then done using a targeted MS(1) based approach, which allowed the identification of a glycation hot spot in the heavy chain complementarity-determining region 3 of the mAb. This targeted approach provided a path forward to developing a structural understanding of the propensity of sites to become glycated on mAbs. Through structural analysis we propose a model in which the number and 3-dimensional distances of carboxylic acid amino acyl residues create a favorable environment for glycation to occur.
Keywords: BA, boronate affinity chromatography; CDR3, complementary-determining region 3; CEX, cation exchange chromatography; CID, collision induced dissociation; CV, coefficient of variation; Da, daltons; EIC, extracted ion chromatogram; HC-CDR3, heavy chain complementary determining region 3; HPLC, high performance liquid chromatography; LC-MS2, liquid chromatography coupled with tandem mass spectrometry; MS1, a mass to charge ratio survey scan; MS2, tandem mass spectrometry - selected ions from MS1 are fragmented and fragment ion mass measured; UPLC, ultrahigh performance liquid chromatography; boronate affinity chromatography; glycation; mAb, monoclonal antibody; structural modeling; targeted mass spectrometry; Å, angstroms.
Figures



Similar articles
-
Hydrogen exchange mass spectrometry reveals protein interfaces and distant dynamic coupling effects during the reversible self-association of an IgG1 monoclonal antibody.MAbs. 2015;7(3):525-39. doi: 10.1080/19420862.2015.1029217. MAbs. 2015. PMID: 25875351 Free PMC article.
-
Characterization of nonenzymatic glycation on a monoclonal antibody.Anal Chem. 2007 Dec 15;79(24):9403-13. doi: 10.1021/ac7017469. Epub 2007 Nov 7. Anal Chem. 2007. PMID: 17985928
-
A study in glycation of a therapeutic recombinant humanized monoclonal antibody: where it is, how it got there, and how it affects charge-based behavior.Anal Biochem. 2008 Feb 15;373(2):179-91. doi: 10.1016/j.ab.2007.09.027. Epub 2007 Sep 29. Anal Biochem. 2008. PMID: 18158144
-
Screening of synthetic PDE-5 inhibitors and their analogues as adulterants: analytical techniques and challenges.J Pharm Biomed Anal. 2014 Jan;87:176-90. doi: 10.1016/j.jpba.2013.04.037. Epub 2013 May 6. J Pharm Biomed Anal. 2014. PMID: 23721687 Review.
-
Detection of oxidized and glycated proteins in clinical samples using mass spectrometry--a user's perspective.Biochim Biophys Acta. 2014 Feb;1840(2):818-29. doi: 10.1016/j.bbagen.2013.03.025. Epub 2013 Apr 2. Biochim Biophys Acta. 2014. PMID: 23558060 Review.
Cited by
-
Isolation and characterization of a monoclonal antibody containing an extra heavy-light chain Fab arm.MAbs. 2018 Apr;10(3):346-353. doi: 10.1080/19420862.2018.1438795. Epub 2018 Mar 20. MAbs. 2018. PMID: 29537936 Free PMC article.
-
The NISTmAb tryptic peptide spectral library for monoclonal antibody characterization.MAbs. 2018 Apr;10(3):354-369. doi: 10.1080/19420862.2018.1436921. Epub 2018 Mar 6. MAbs. 2018. PMID: 29425077 Free PMC article.
-
Glycation of antibodies: Modification, methods and potential effects on biological functions.MAbs. 2017 May/Jun;9(4):586-594. doi: 10.1080/19420862.2017.1300214. Epub 2017 Mar 8. MAbs. 2017. PMID: 28272973 Free PMC article. Review.
-
Resolving the micro-heterogeneity and structural integrity of monoclonal antibodies by hybrid mass spectrometric approaches.MAbs. 2017 May/Jun;9(4):638-645. doi: 10.1080/19420862.2017.1290033. Epub 2017 Feb 14. MAbs. 2017. PMID: 28281873 Free PMC article.
-
In silico prediction of post-translational modifications in therapeutic antibodies.MAbs. 2022 Jan-Dec;14(1):2023938. doi: 10.1080/19420862.2021.2023938. MAbs. 2022. PMID: 35040751 Free PMC article. Review.
References
-
- Maillard L. Action des acides amines sure les sucres; formation des melanoidines par voie methodique. Comptes Rendus Hebdomadaires Des Seances De L'academie Des Sciences 1912; 154:66-8
-
- Priego Capote F, Sanchez JC. Strategies for proteomic analysis of non-enzymatically glycated proteins. Mass Spectr Rev 2009; 28:135-46; PMID:18949816; http://dx.doi.org/10.1002/mas.20187 - DOI - PubMed
-
- Baynes JW, Watkins NG, Fisher CI, Hull CJ, Patrick JS, Ahmed MU, Dunn JA, Thorpe SR. The Amadori product on protein: structure and reactions. Prog Clin Biol Res 1989; 304:43-67; PMID:2675036 - PubMed
-
- Singh R, Barden A, Mori T, Beilin L. Advanced glycation end-products: a review. Diabetologia 2001; 44:129-46; PMID:11270668; http://dx.doi.org/10.1007/s001250051591 - DOI - PubMed
-
- Frye EB, Degenhardt TP, Thorpe SR, Baynes JW. Role of the Maillard reaction in aging of tissue proteins. Advanced glycation end product-dependent increase in imidazolium cross-links in human lens proteins. J Biol Chem 1998; 273:18714-9; PMID:9668043; http://dx.doi.org/10.1074/jbc.273.30.18714 - DOI - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
