Prmt5 is a regulator of muscle stem cell expansion in adult mice
- PMID: 26028225
- PMCID: PMC4458870
- DOI: 10.1038/ncomms8140
Prmt5 is a regulator of muscle stem cell expansion in adult mice
Abstract
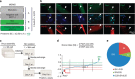
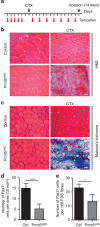
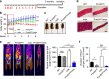
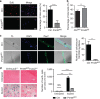
Skeletal muscle stem cells (MuSC), also called satellite cells, are indispensable for maintenance and regeneration of adult skeletal muscles. Yet, a comprehensive picture of the regulatory events controlling the fate of MuSC is missing. Here, we determine the proteome of MuSC to design a loss-of-function screen, and identify 120 genes important for MuSC function including the arginine methyltransferase Prmt5. MuSC-specific inactivation of Prmt5 in adult mice prevents expansion of MuSC, abolishes long-term MuSC maintenance and abrogates skeletal muscle regeneration. Interestingly, Prmt5 is dispensable for proliferation and differentiation of Pax7(+) myogenic progenitor cells during mouse embryonic development, indicating significant differences between embryonic and adult myogenesis. Mechanistic studies reveal that Prmt5 controls proliferation of adult MuSC by direct epigenetic silencing of the cell cycle inhibitor p21. We reason that Prmt5 generates a poised state that keeps MuSC in a standby mode, thus allowing rapid MuSC amplification under disease conditions.
Figures
Similar articles
-
PRMT7 Preserves Satellite Cell Regenerative Capacity.Cell Rep. 2016 Feb 16;14(6):1528-1539. doi: 10.1016/j.celrep.2016.01.022. Epub 2016 Feb 4. Cell Rep. 2016. PMID: 26854227
-
Concise Review: Epigenetic Regulation of Myogenesis in Health and Disease.Stem Cells Transl Med. 2016 Mar;5(3):282-90. doi: 10.5966/sctm.2015-0266. Epub 2016 Jan 21. Stem Cells Transl Med. 2016. PMID: 26798058 Free PMC article. Review.
-
MyoD Over-Expression Rescues GST-bFGF Repressed Myogenesis.Int J Mol Sci. 2024 Apr 13;25(8):4308. doi: 10.3390/ijms25084308. Int J Mol Sci. 2024. PMID: 38673893 Free PMC article.
-
microRNA-1 and microRNA-206 regulate skeletal muscle satellite cell proliferation and differentiation by repressing Pax7.J Cell Biol. 2010 Sep 6;190(5):867-79. doi: 10.1083/jcb.200911036. J Cell Biol. 2010. PMID: 20819939 Free PMC article.
-
Regulation of skeletal muscle stem cells through epigenetic mechanisms.Toxicol Mech Methods. 2011 May;21(4):334-42. doi: 10.3109/15376516.2011.557873. Toxicol Mech Methods. 2011. PMID: 21495871 Review.
Cited by
-
Transcriptional regulation by methyltransferases and their role in the heart: highlighting novel emerging functionality.Am J Physiol Heart Circ Physiol. 2020 Oct 1;319(4):H847-H865. doi: 10.1152/ajpheart.00382.2020. Epub 2020 Aug 21. Am J Physiol Heart Circ Physiol. 2020. PMID: 32822544 Free PMC article. Review.
-
Linc-MYH configures INO80 to regulate muscle stem cell numbers and skeletal muscle hypertrophy.EMBO J. 2020 Nov 16;39(22):e105098. doi: 10.15252/embj.2020105098. Epub 2020 Sep 22. EMBO J. 2020. PMID: 32960481 Free PMC article.
-
Protein Arginine Methyltransferases in Neuromuscular Function and Diseases.Cells. 2022 Jan 21;11(3):364. doi: 10.3390/cells11030364. Cells. 2022. PMID: 35159176 Free PMC article. Review.
-
Transcriptomic Profiling of Skeletal Muscle Reveals Candidate Genes Influencing Muscle Growth and Associated Lipid Composition in Portuguese Local Pig Breeds.Animals (Basel). 2021 May 16;11(5):1423. doi: 10.3390/ani11051423. Animals (Basel). 2021. PMID: 34065673 Free PMC article.
-
Fibroblast-specific PRMT5 deficiency suppresses cardiac fibrosis and left ventricular dysfunction in male mice.Nat Commun. 2024 Mar 19;15(1):2472. doi: 10.1038/s41467-024-46711-z. Nat Commun. 2024. PMID: 38503742 Free PMC article.
References
-
- Wilson A. et al. Hematopoietic stem cells reversibly switch from dormancy to self-renewal during homeostasis and repair. Cell 135, 1118–1129 (2008). - PubMed
-
- Seale P. et al. Pax7 is required for the specification of myogenic satellite cells. Cell 102, 777–786 (2000). - PubMed
-
- Relaix F., Rocancourt D., Mansouri A. & Buckingham M. A Pax3/Pax7-dependent population of skeletal muscle progenitor cells. Nature 435, 948–953 (2005). - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases

