Mechanistic Analysis of Activation of the Innate Immune Sensor PKR by Bacterial RNA
- PMID: 26026708
- PMCID: PMC4624505
- DOI: 10.1016/j.jmb.2015.05.018
Mechanistic Analysis of Activation of the Innate Immune Sensor PKR by Bacterial RNA
Abstract
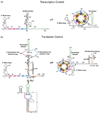
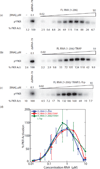
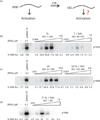
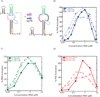
The protein kinase PKR (protein kinase R) is a sensor in innate immunity. PKR autophosphorylates in the presence of double-stranded RNA enabling it to phosphorylate its substrate, eIF2α (eukaryotic initiation factor 2α), halting cellular translation. Classical activators of PKR are long viral double-stranded RNAs, but recently, PKR has been found to be activated by bacterial RNA. However, the features of bacterial RNA that activate PKR are unknown. We studied the Bacillus subtilis trp 5'-UTR (untranslated region), which is an indirect riboswitch with secondary and tertiary RNA structures that regulate gene function. Additionally, the trp 5'-UTR binds a protein, TRAP (tryptophan RNA-binding attenuation protein), which recognizes l-tryptophan. We present the first evidence that multiple structural features in this RNA, which are typical of bacterial RNAs, activate PKR in TRAP-free and TRAP/l-Trp-bound forms. Segments from the 5'-UTR, including the terminator 5'-stem-loop and Shine-Dalgarno blocking hairpins, demonstrated 5'-triphosphate and flanking RNA tail dependence on PKR activation. Disruption of long-distance tertiary interactions in the 5'-UTR led to partial loss in activation, consistent with highly base-paired regions in bacterial RNA activating PKR. One physiological change a bacterial RNA would face in a human cell is a decrease in the concentration of free magnesium. Upon lowering the magnesium concentration to human physiological conditions of 0.5mM, the trp 5'-UTR continued to activate PKR potently. Moreover, total RNA from Escherichia coli, depleted of rRNA, also activated PKR under these ionic conditions. This study demonstrates that PKR can signal the presence of bacterial RNAs under physiological ionic conditions and offers a potential explanation for the apparent absence of riboswitches in the human genome.
Keywords: PKR; RNA folding; bacterial RNA; innate immunity; riboswitch.
Copyright © 2015 Elsevier Ltd. All rights reserved.
Figures


Similar articles
-
Discriminating Self and Non-Self by RNA: Roles for RNA Structure, Misfolding, and Modification in Regulating the Innate Immune Sensor PKR.Acc Chem Res. 2016 Jun 21;49(6):1242-9. doi: 10.1021/acs.accounts.6b00151. Epub 2016 Jun 8. Acc Chem Res. 2016. PMID: 27269119 Free PMC article. Review.
-
Mechanistic characterization of the 5'-triphosphate-dependent activation of PKR: lack of 5'-end nucleobase specificity, evidence for a distinct triphosphate binding site, and a critical role for the dsRBD.RNA. 2012 Oct;18(10):1862-74. doi: 10.1261/rna.034520.112. Epub 2012 Aug 21. RNA. 2012. PMID: 22912486 Free PMC article.
-
A Mg2+-dependent RNA tertiary structure forms in the Bacillus subtilis trp operon leader transcript and appears to interfere with trpE translation control by inhibiting TRAP binding.J Mol Biol. 2003 Sep 19;332(3):555-74. doi: 10.1016/s0022-2836(03)00969-0. J Mol Biol. 2003. PMID: 12963367
-
trp RNA-binding attenuation protein-5' stem-loop RNA interaction is required for proper transcription attenuation control of the Bacillus subtilis trpEDCFBA operon.J Bacteriol. 2000 Apr;182(7):1819-27. doi: 10.1128/JB.182.7.1819-1827.2000. J Bacteriol. 2000. PMID: 10714985 Free PMC article.
-
Regulation of transcription attenuation and translation initiation by allosteric control of an RNA-binding protein: the Bacillus subtilis TRAP protein.Curr Opin Microbiol. 2004 Apr;7(2):132-9. doi: 10.1016/j.mib.2004.02.003. Curr Opin Microbiol. 2004. PMID: 15063849 Review.
Cited by
-
Discriminating Self and Non-Self by RNA: Roles for RNA Structure, Misfolding, and Modification in Regulating the Innate Immune Sensor PKR.Acc Chem Res. 2016 Jun 21;49(6):1242-9. doi: 10.1021/acs.accounts.6b00151. Epub 2016 Jun 8. Acc Chem Res. 2016. PMID: 27269119 Free PMC article. Review.
-
SIDT2 RNA Transporter Promotes Lung and Gastrointestinal Tumor Development.iScience. 2019 Oct 25;20:14-24. doi: 10.1016/j.isci.2019.09.009. Epub 2019 Sep 10. iScience. 2019. PMID: 31546103 Free PMC article.
-
Protein Kinase R in Bacterial Infections: Friend or Foe?Front Immunol. 2021 Jul 8;12:702142. doi: 10.3389/fimmu.2021.702142. eCollection 2021. Front Immunol. 2021. PMID: 34305942 Free PMC article. Review.
-
Cytoplasmic RNA Sensor Pathways and Nitazoxanide Broadly Inhibit Intracellular Mycobacterium tuberculosis Growth.iScience. 2019 Dec 20;22:299-313. doi: 10.1016/j.isci.2019.11.001. Epub 2019 Nov 6. iScience. 2019. PMID: 31805434 Free PMC article.
-
Bridging the gap between in vitro and in vivo RNA folding.Q Rev Biophys. 2016 Jan;49:e10. doi: 10.1017/S003358351600007X. Epub 2016 Jun 24. Q Rev Biophys. 2016. PMID: 27658939 Free PMC article.
References
-
- Balachandran S, Roberts PC, Brown LE, Truong H, Pattnaik AK, Archer DR, Barber GN. Essential role for the dsRNA-dependent protein kinase PKR in innate immunity to viral infection. Immunity. 2000;13:129–141. - PubMed
-
- Green SR, Mathews MB. Two RNA-binding motifs in the double-stranded RNA-activated protein kinase, DAI. Genes Dev. 1992;6:2478–2490. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

