The V1V2 Region of HIV-1 gp120 Forms a Five-Stranded Beta Barrel
- PMID: 26018158
- PMCID: PMC4505664
- DOI: 10.1128/JVI.00754-15
The V1V2 Region of HIV-1 gp120 Forms a Five-Stranded Beta Barrel
Abstract
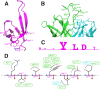
The region consisting of the first and second variable regions (V1V2) of gp120 plays vital roles in the functioning of the HIV-1 envelope (Env). V1V2, which harbors multiple glycans and is highly sequence diverse, is located at the Env apex and stabilizes the trimeric gp120 spike on the virion surface. It shields V3 and the coreceptor binding sites in the prefusion state and exposes them upon CD4 binding. Data from the RV144 human HIV-1 vaccine trial suggested that antibody responses targeting the V1V2 region inversely correlated with the risk of infection; thus, understanding the antigenic structure of V1V2 can contribute to vaccine design. We have determined a crystal structure of a V1V2 scaffold molecule (V1V2ZM109-1FD6) in complex with 830A, a human monoclonal antibody that recognizes a V1V2 epitope overlapping the integrin-binding motif in V2. The structure revealed that V1V2 assumes a five-stranded beta barrel structure with the region of the integrin-binding site (amino acids [aa] 179 to 181) included in a "kink" followed by an extra beta strand. The complete barrel structure naturally presents the glycans on its outer surface and packs into its core conserved hydrophobic residues, including the Ile at position 181 which was highly correlated with vaccine efficacy in RV144. The epitope of monoclonal antibody 830A is discontinuous and composed of three segments: (i) Thr175, Tyr177, Leu179, and Asp180 at the kink overlapping the integrin-binding site; (ii) Arg153 and Val154 in V1; and (iii) Ile194 at the C terminus of V2. This report thus provides the atomic details of the immunogenic "V2i epitope."
Importance: Data from the RV144 phase III clinical trial suggested that the presence of antibodies to the first and second variable regions (V1V2) of gp120 was associated with the modest protection afforded by the vaccine. V1V2 is a highly variable and immunogenic region of HIV-1 surface glycoprotein gp120, and structural information about this region and its antigenic landscape will be crucial in the design of an effective HIV-1 vaccine. We have determined a crystal structure of V1V2 in complex with human MAb 830A and have shown that MAb 830A recognizes a region overlapping the α4β7 integrin-binding site. We also showed that V1V2 forms a 5-stranded beta barrel, an elegant structure allowing sequence variations in the strand-connecting loops while preserving a conserved core.
Copyright © 2015, American Society for Microbiology. All Rights Reserved.
Figures



Similar articles
-
Rationally Targeted Mutations at the V1V2 Domain of the HIV-1 Envelope to Augment Virus Neutralization by Anti-V1V2 Monoclonal Antibodies.PLoS One. 2015 Oct 22;10(10):e0141233. doi: 10.1371/journal.pone.0141233. eCollection 2015. PLoS One. 2015. PMID: 26491873 Free PMC article.
-
Functional implications of the binding mode of a human conformation-dependent V2 monoclonal antibody against HIV.J Virol. 2014 Apr;88(8):4100-12. doi: 10.1128/JVI.03153-13. Epub 2014 Jan 29. J Virol. 2014. PMID: 24478429 Free PMC article.
-
Rationally Designed Immunogens Targeting HIV-1 gp120 V1V2 Induce Distinct Conformation-Specific Antibody Responses in Rabbits.J Virol. 2016 Nov 28;90(24):11007-11019. doi: 10.1128/JVI.01409-16. Print 2016 Dec 15. J Virol. 2016. PMID: 27707920 Free PMC article.
-
Vaccine-induced V1V2-specific antibodies control and or protect against infection with HIV, SIV and SHIV.Curr Opin HIV AIDS. 2019 Jul;14(4):309-317. doi: 10.1097/COH.0000000000000551. Curr Opin HIV AIDS. 2019. PMID: 30994501 Free PMC article. Review.
-
The HIV-1 gp120 V1V2 loop: structure, function and importance for vaccine development.Expert Rev Vaccines. 2014 Dec;13(12):1489-500. doi: 10.1586/14760584.2014.951335. Epub 2014 Aug 28. Expert Rev Vaccines. 2014. PMID: 25163695 Review.
Cited by
-
Mucosal Delivery of HIV-1 Glycoprotein Vaccine Candidate Enabled by Short Carbon Nanotubes.Part Part Syst Charact. 2022 May;39(5):2200011. doi: 10.1002/ppsc.202200011. Epub 2022 Mar 7. Part Part Syst Charact. 2022. PMID: 36186663 Free PMC article.
-
V2-Specific Antibodies in HIV-1 Vaccine Research and Natural Infection: Controllers or Surrogate Markers.Vaccines (Basel). 2019 Aug 6;7(3):82. doi: 10.3390/vaccines7030082. Vaccines (Basel). 2019. PMID: 31390725 Free PMC article. Review.
-
Broadly neutralizing antibody epitopes on HIV-1 particles are exposed after virus interaction with host cells.J Virol. 2023 Sep 28;97(9):e0071023. doi: 10.1128/jvi.00710-23. Epub 2023 Sep 8. J Virol. 2023. PMID: 37681958 Free PMC article.
-
Non-neutralizing Antibodies Targeting the V1V2 Domain of HIV Exhibit Strong Antibody-Dependent Cell-mediated Cytotoxic Activity.Sci Rep. 2017 Oct 4;7(1):12655. doi: 10.1038/s41598-017-12883-6. Sci Rep. 2017. PMID: 28978939 Free PMC article.
-
Asymmetric recognition of HIV-1 Envelope trimer by V1V2 loop-targeting antibodies.Elife. 2017 May 26;6:e27389. doi: 10.7554/eLife.27389. Elife. 2017. PMID: 28548638 Free PMC article.
References
-
- Leonard CK, Spellman MW, Riddle L, Harris RJ, Thomas JN, Gregory TJ. 1990. Assignment of intrachain disulfide bonds and characterization of potential glycosylation sites of the type 1 recombinant human immunodeficiency virus envelope glycoprotein (gp120) expressed in Chinese hamster ovary cells. J Biol Chem 265:10373–10382. - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases
Research Materials
Miscellaneous

