The fitness consequences of aneuploidy are driven by condition-dependent gene effects
- PMID: 26011532
- PMCID: PMC4444335
- DOI: 10.1371/journal.pbio.1002155
The fitness consequences of aneuploidy are driven by condition-dependent gene effects
Abstract
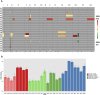
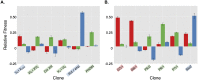
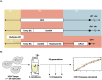
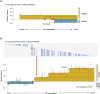
Aneuploidy is a hallmark of tumor cells, and yet the precise relationship between aneuploidy and a cell's proliferative ability, or cellular fitness, has remained elusive. In this study, we have combined a detailed analysis of aneuploid clones isolated from laboratory-evolved populations of Saccharomyces cerevisiae with a systematic, genome-wide screen for the fitness effects of telomeric amplifications to address the relationship between aneuploidy and cellular fitness. We found that aneuploid clones rise to high population frequencies in nutrient-limited evolution experiments and show increased fitness relative to wild type. Direct competition experiments confirmed that three out of four aneuploid events isolated from evolved populations were themselves sufficient to improve fitness. To expand the scope beyond this small number of exemplars, we created a genome-wide collection of >1,800 diploid yeast strains, each containing a different telomeric amplicon (Tamp), ranging in size from 0.4 to 1,000 kb. Using pooled competition experiments in nutrient-limited chemostats followed by high-throughput sequencing of strain-identifying barcodes, we determined the fitness effects of these >1,800 Tamps under three different conditions. Our data revealed that the fitness landscape explored by telomeric amplifications is much broader than that explored by single-gene amplifications. As also observed in the evolved clones, we found the fitness effects of most Tamps to be condition specific, with a minority showing common effects in all three conditions. By integrating our data with previous work that examined the fitness effects of single-gene amplifications genome-wide, we found that a small number of genes within each Tamp are centrally responsible for each Tamp's fitness effects. Our genome-wide Tamp screen confirmed that telomeric amplifications identified in laboratory-evolved populations generally increased fitness. Our results show that Tamps are mutations that produce large, typically condition-dependent changes in fitness that are important drivers of increased fitness in asexually evolving populations.
Conflict of interest statement
The authors have declared that no competing interests exist.
Figures


Similar articles
-
Condition-dependent fitness effects of large synthetic chromosome amplifications.bioRxiv [Preprint]. 2023 Jun 9:2023.06.08.544269. doi: 10.1101/2023.06.08.544269. bioRxiv. 2023. PMID: 37333112 Free PMC article. Preprint.
-
Chromosome-Specific and Global Effects of Aneuploidy in Saccharomyces cerevisiae.Genetics. 2016 Apr;202(4):1395-409. doi: 10.1534/genetics.115.185660. Epub 2016 Feb 2. Genetics. 2016. PMID: 26837754 Free PMC article.
-
High-Throughput Identification of Adaptive Mutations in Experimentally Evolved Yeast Populations.PLoS Genet. 2016 Oct 11;12(10):e1006339. doi: 10.1371/journal.pgen.1006339. eCollection 2016 Oct. PLoS Genet. 2016. PMID: 27727276 Free PMC article.
-
Utilizing Saccharomyces cerevisiae to identify aneuploidy and cancer susceptibility genes.Methods Mol Biol. 2010;653:73-85. doi: 10.1007/978-1-60761-759-4_5. Methods Mol Biol. 2010. PMID: 20721738 Review.
-
The aneuploidy paradox: costs and benefits of an incorrect karyotype.Trends Genet. 2011 Nov;27(11):446-53. doi: 10.1016/j.tig.2011.07.003. Epub 2011 Aug 26. Trends Genet. 2011. PMID: 21872963 Free PMC article. Review.
Cited by
-
Highly parallelized laboratory evolution of wine yeasts for enhanced metabolic phenotypes.Mol Syst Biol. 2024 Oct;20(10):1109-1133. doi: 10.1038/s44320-024-00059-0. Epub 2024 Aug 22. Mol Syst Biol. 2024. PMID: 39174863 Free PMC article.
-
Ploidy dynamics and evolvability in fungi.Philos Trans R Soc Lond B Biol Sci. 2016 Dec 5;371(1709):20150461. doi: 10.1098/rstb.2015.0461. Philos Trans R Soc Lond B Biol Sci. 2016. PMID: 28080987 Free PMC article. Review.
-
A fitness trade-off explains the early fate of yeast aneuploids with chromosome gains.Proc Natl Acad Sci U S A. 2023 Apr 11;120(15):e2211687120. doi: 10.1073/pnas.2211687120. Epub 2023 Apr 5. Proc Natl Acad Sci U S A. 2023. PMID: 37018197 Free PMC article.
-
Multilevel gene expression changes in lineages containing adaptive copy number variants.bioRxiv [Preprint]. 2024 Jul 9:2023.10.20.563336. doi: 10.1101/2023.10.20.563336. bioRxiv. 2024. Update in: Mol Biol Evol. 2025 Jan 23:msaf005. doi: 10.1093/molbev/msaf005 PMID: 37961325 Free PMC article. Updated. Preprint.
-
Exploring genetic suppression interactions on a global scale.Science. 2016 Nov 4;354(6312):aag0839. doi: 10.1126/science.aag0839. Science. 2016. PMID: 27811238 Free PMC article.
References
-
- Boveri T (1902) Über mehrpolige Mitosen als Mittel zur Analyse des Zellkerns. Verhandlungen der physikalisch-medicinischen Gesellschaft zu Würzburg 35: 67–90. http://publikationen.ub.uni-frankfurt.de/frontdoor/index/index/docId/15991.
-
- Weaver BAA, Silk AD, Montagna C, Verdier-Pinard P, Cleveland DW (2007) Aneuploidy acts both oncogenically and as a tumor suppressor. Cancer Cell 11: 25–36. - PubMed
-
- Torres E, Sokolsky T, Tucker C, Chan L, Boselli M, et al. (2007) Effects of aneuploidy on cellular physiology and cell division in haploid yeast. Science 317: 916 - PubMed
Publication types
MeSH terms
Associated data
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials

