Transmission dynamics of pandemic influenza A(H1N1)pdm09 virus in humans and swine in backyard farms in Tumbes, Peru
- PMID: 26011186
- PMCID: PMC4687498
- DOI: 10.1111/irv.12329
Transmission dynamics of pandemic influenza A(H1N1)pdm09 virus in humans and swine in backyard farms in Tumbes, Peru
Abstract
Objectives: We aimed to determine the frequency of pH1N1 transmission between humans and swine on backyard farms in Tumbes, Peru.
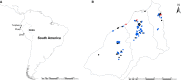
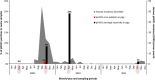
Design: Two-year serial cross-sectional study comprising four sampling periods: March 2009 (pre-pandemic), October 2009 (peak of the pandemic in Peru), April 2010 (1st post-pandemic period), and October 2011 (2nd post-pandemic period).
Sample: Backyard swine serum, tracheal swabs, and lung sample were collected during each sampling period.
Main outcome measures: We assessed current and past pH1N1 infection in swine through serological testing, virus culture, and RT-PCR and compared the results with human incidence data from a population-based active surveillance cohort study in Peru.
Results: Among 1303 swine sampled, the antibody prevalence to pH1N1 was 0% pre-pandemic, 8% at the peak of the human pandemic (October 2009), and 24% in April 2010 and 1% in October 2011 (post-pandemic sampling periods). Trends in swine seropositivity paralleled those seen in humans in Tumbes. The pH1N1 virus was isolated from three pigs during the peak of the pandemic. Phylogenetic analysis revealed that these viruses likely represent two separate human-to-swine transmission events in backyard farm settings.
Conclusions: Our findings suggest that human-to-swine pH1N1 transmission occurred during the pandemic among backyard farms in Peru, emphasizing the importance of interspecies transmission in backyard pig populations. Continued surveillance for influenza viruses in backyard farms is warranted.
Keywords: Antibodies; backyard pig farms; human-animal transmission; influenza.
© 2015 The Authors. Influenza and Other Respiratory Viruses Published by John Wiley & Sons Ltd.
Figures

Similar articles
-
Serological evidence of pig-to-human influenza virus transmission on Thai swine farms.Vet Microbiol. 2011 Mar 24;148(2-4):413-8. doi: 10.1016/j.vetmic.2010.09.019. Epub 2010 Sep 29. Vet Microbiol. 2011. PMID: 20965670
-
Continual Reintroduction of Human Pandemic H1N1 Influenza A Viruses into Swine in the United States, 2009 to 2014.J Virol. 2015 Jun;89(12):6218-26. doi: 10.1128/JVI.00459-15. Epub 2015 Apr 1. J Virol. 2015. PMID: 25833052 Free PMC article.
-
Bidirectional Human-Swine Transmission of Seasonal Influenza A(H1N1)pdm09 Virus in Pig Herd, France, 2018.Emerg Infect Dis. 2019 Oct;25(10):1940-1943. doi: 10.3201/eid2510.190068. Emerg Infect Dis. 2019. PMID: 31538914 Free PMC article.
-
Reverse zoonosis of influenza to swine: new perspectives on the human-animal interface.Trends Microbiol. 2015 Mar;23(3):142-53. doi: 10.1016/j.tim.2014.12.002. Epub 2015 Jan 4. Trends Microbiol. 2015. PMID: 25564096 Free PMC article. Review.
-
[An overview on swine influenza viruses].Bing Du Xue Bao. 2013 May;29(3):330-6. Bing Du Xue Bao. 2013. PMID: 23905479 Review. Chinese.
Cited by
-
Risk factors and spatial relative risk assessment for influenza A virus in poultry and swine in backyard production systems of central Chile.Vet Med Sci. 2020 Aug;6(3):518-526. doi: 10.1002/vms3.254. Epub 2020 Feb 21. Vet Med Sci. 2020. PMID: 32086880 Free PMC article.
-
Origins of the 2009 H1N1 influenza pandemic in swine in Mexico.Elife. 2016 Jun 28;5:e16777. doi: 10.7554/eLife.16777. Elife. 2016. PMID: 27350259 Free PMC article.
-
Novel Human-like Influenza A Viruses Circulate in Swine in Mexico and Chile.PLoS Curr. 2015 Aug 13;7:ecurrents.outbreaks.c8b3207c9bad98474eca3013fa933ca6. doi: 10.1371/currents.outbreaks.c8b3207c9bad98474eca3013fa933ca6. PLoS Curr. 2015. PMID: 26345598 Free PMC article.
-
Improving Current Knowledge on Seroprevalence and Genetic Characterization of Swine Influenza Virus in Croatian Pig Farms: A Retrospective Study.Pathogens. 2021 Nov 22;10(11):1527. doi: 10.3390/pathogens10111527. Pathogens. 2021. PMID: 34832682 Free PMC article.
-
A Systematic Review Analyzing the Prevalence and Circulation of Influenza Viruses in Swine Population Worldwide.Pathogens. 2020 May 8;9(5):355. doi: 10.3390/pathogens9050355. Pathogens. 2020. PMID: 32397138 Free PMC article. Review.
References
Publication types
MeSH terms
Associated data
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Miscellaneous

