The mitochondrial uncoupler DNP triggers brain cell mTOR signaling network reprogramming and CREB pathway up-regulation
- PMID: 26010875
- PMCID: PMC4516713
- DOI: 10.1111/jnc.13176
The mitochondrial uncoupler DNP triggers brain cell mTOR signaling network reprogramming and CREB pathway up-regulation
Abstract
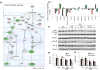
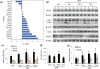
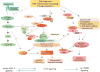
Mitochondrial metabolism is highly responsive to nutrient availability and ongoing activity in neuronal circuits. The molecular mechanisms by which brain cells respond to an increase in cellular energy expenditure are largely unknown. Mild mitochondrial uncoupling enhances cellular energy expenditure in mitochondria and can be induced with 2,4-dinitrophenol (DNP), a proton ionophore previously used for weight loss. We found that DNP treatment reduces mitochondrial membrane potential, increases intracellular Ca(2+) levels and reduces oxidative stress in cerebral cortical neurons. Gene expression profiling of the cerebral cortex of DNP-treated mice revealed reprogramming of signaling cascades that included suppression of the mammalian target of rapamycin (mTOR) and insulin--PI3K - MAPK pathways, and up-regulation of tuberous sclerosis complex 2, a negative regulator of mTOR. Genes encoding proteins involved in autophagy processes were up-regulated in response to DNP. CREB (cAMP-response element-binding protein) signaling, Arc and brain-derived neurotrophic factor, which play important roles in synaptic plasticity and adaptive cellular stress responses, were up-regulated in response to DNP, and DNP-treated mice exhibited improved performance in a test of learning and memory. Immunoblot analysis verified that key DNP-induced changes in gene expression resulted in corresponding changes at the protein level. Our findings suggest that mild mitochondrial uncoupling triggers an integrated signaling response in brain cells characterized by reprogramming of mTOR and insulin signaling, and up-regulation of pathways involved in adaptive stress responses, molecular waste disposal, and synaptic plasticity. Physiological bioenergetic challenges such as exercise and fasting can enhance neuroplasticity and protect neurons against injury and neurodegeneration. Here, we show that the mitochondrial uncoupling agent 2,4-dinitrophenol (DNP) elicits adaptive signaling responses in the cerebral cortex involving activation of Ca(2+) -CREB and autophagy pathways, and inhibition of mTOR and insulin signaling pathways. The molecular reprogramming induced by DNP, which is similar to that of exercise and fasting, is associated with improved learning and memory, suggesting potential therapeutic applications for DNP.
Keywords: 2,4-dinitrophenol; BDNF; CREB; autophagy; insulin signaling; mTOR.
© 2015 International Society for Neurochemistry.
Conflict of interest statement
The authors declare no competing interests.
Figures



Similar articles
-
DNP, mitochondrial uncoupling, and neuroprotection: A little dab'll do ya.Alzheimers Dement. 2017 May;13(5):582-591. doi: 10.1016/j.jalz.2016.08.001. Epub 2016 Sep 4. Alzheimers Dement. 2017. PMID: 27599210 Free PMC article.
-
Mitochondrial uncoupling reduces exercise capacity despite several skeletal muscle metabolic adaptations.J Appl Physiol (1985). 2014 Feb 15;116(4):364-75. doi: 10.1152/japplphysiol.01177.2013. Epub 2013 Dec 12. J Appl Physiol (1985). 2014. PMID: 24336883
-
Protective effects of a green tea polyphenol, epigallocatechin-3-gallate, against sevoflurane-induced neuronal apoptosis involve regulation of CREB/BDNF/TrkB and PI3K/Akt/mTOR signalling pathways in neonatal mice.Can J Physiol Pharmacol. 2017 Dec;95(12):1396-1405. doi: 10.1139/cjpp-2016-0333. Epub 2017 Jul 5. Can J Physiol Pharmacol. 2017. PMID: 28679060
-
Adaptive responses of neuronal mitochondria to bioenergetic challenges: Roles in neuroplasticity and disease resistance.Free Radic Biol Med. 2017 Jan;102:203-216. doi: 10.1016/j.freeradbiomed.2016.11.045. Epub 2016 Nov 29. Free Radic Biol Med. 2017. PMID: 27908782 Free PMC article. Review.
-
Targeted mitochondrial uncoupling beyond UCP1 - The fine line between death and metabolic health.Biochimie. 2017 Mar;134:77-85. doi: 10.1016/j.biochi.2016.11.013. Epub 2016 Dec 2. Biochimie. 2017. PMID: 27916644 Review.
Cited by
-
Is cancer a severe delayed hypersensitivity reaction and histamine a blueprint?Clin Transl Med. 2016 Dec;5(1):35. doi: 10.1186/s40169-016-0108-3. Epub 2016 Aug 23. Clin Transl Med. 2016. PMID: 27558401 Free PMC article.
-
The role of mitochondrial uncoupling in the regulation of mitostasis after traumatic brain injury.Neurochem Int. 2024 Mar;174:105680. doi: 10.1016/j.neuint.2024.105680. Epub 2024 Feb 3. Neurochem Int. 2024. PMID: 38311216
-
Current Advances in Mitochondrial Targeted Interventions in Alzheimer's Disease.Biomedicines. 2023 Aug 22;11(9):2331. doi: 10.3390/biomedicines11092331. Biomedicines. 2023. PMID: 37760774 Free PMC article. Review.
-
Hallmarks of Brain Aging: Adaptive and Pathological Modification by Metabolic States.Cell Metab. 2018 Jun 5;27(6):1176-1199. doi: 10.1016/j.cmet.2018.05.011. Cell Metab. 2018. PMID: 29874566 Free PMC article. Review.
-
Mitochondrial uncoupling in the melanocortin system differentially regulates NPY and POMC neurons to promote weight-loss.Mol Metab. 2017 Oct;6(10):1103-1112. doi: 10.1016/j.molmet.2017.07.002. Epub 2017 Jul 8. Mol Metab. 2017. PMID: 29031712 Free PMC article.
References
-
- Andrews ZB, Diano S, Horvath TL. Mitochondrial uncoupling proteins in the CNS: in support of function and survival. Nat Rev Neurosci. 2005;6:829–40. - PubMed
-
- Arancibia S, Rage F, Givaloi L, Tapia-Arancibia L. Neurotrophins in neuroendocrine control: brain derived-neurotrophic factor (BDNF) and somatostatin involvement in the stress response and reproductive physiology. Ann Rev Biomed Sciences. 2003;5:5–28.
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

