Spring loading a pre-cleavage intermediate for hairpin telomere formation
- PMID: 26007659
- PMCID: PMC4499125
- DOI: 10.1093/nar/gkv497
Spring loading a pre-cleavage intermediate for hairpin telomere formation
Abstract
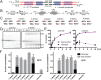
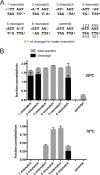
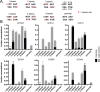
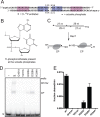
The Borrelia telomere resolvase, ResT, forms the unusual hairpin telomeres of the linear Borrelia replicons in a process referred to as telomere resolution. Telomere resolution is a DNA cleavage and rejoining reaction that proceeds from a replicated telomere intermediate in a reaction with mechanistic similarities to that catalyzed by type IB topoisomerases. Previous reports have implicated the hairpin-binding module, at the end of the N-terminal domain of ResT, in distorting the DNA between the scissile phosphates so as to promote DNA cleavage and hairpin formation by the catalytic domain. We report that unwinding the DNA between the scissile phosphates, prior to DNA cleavage, is a key cold-sensitive step in telomere resolution. Through the analysis of ResT mutants, rescued by substrate modifications that mimic DNA unwinding between the cleavage sites, we show that formation and/or stabilization of an underwound pre-cleavage intermediate depends upon cooperation of the hairpin-binding module and catalytic domain. The phenotype of the mutants argues that the pre-cleavage intermediate promotes strand ejection to favor the forward reaction and that subsequent hairpin capture is a reversible reaction step. These reaction features are proposed to promote hairpin formation over strand resealing while allowing reversal back to substrate of aborted reactions.
© The Author(s) 2015. Published by Oxford University Press on behalf of Nucleic Acids Research.
Figures

Similar articles
-
The telomere resolvase, TelA, utilizes an underwound pre-cleavage intermediate to promote hairpin telomere formation.PLoS One. 2023 Nov 29;18(11):e0294732. doi: 10.1371/journal.pone.0294732. eCollection 2023. PLoS One. 2023. PMID: 38019799 Free PMC article.
-
Uncoupling the chemical steps of telomere resolution by ResT.J Biol Chem. 2005 Jul 22;280(29):26788-95. doi: 10.1074/jbc.M504530200. Epub 2005 May 23. J Biol Chem. 2005. PMID: 15917226
-
Telomere resolution by Borrelia burgdorferi ResT through the collaborative efforts of tethered DNA binding domains.Mol Microbiol. 2007 May;64(3):580-90. doi: 10.1111/j.1365-2958.2007.05691.x. Mol Microbiol. 2007. PMID: 17462009
-
Split target specificity of ResT: a design for protein delivery, site selectivity and regulation of enzyme activity?Mol Microbiol. 2007 May;64(3):575-9. doi: 10.1111/j.1365-2958.2007.05700.x. Mol Microbiol. 2007. PMID: 17462008 Review.
-
Hairpin telomeres and genome plasticity in Borrelia: all mixed up in the end.Mol Microbiol. 2005 Nov;58(3):625-35. doi: 10.1111/j.1365-2958.2005.04872.x. Mol Microbiol. 2005. PMID: 16238614 Review.
Cited by
-
The telomere resolvase, TelA, utilizes an underwound pre-cleavage intermediate to promote hairpin telomere formation.PLoS One. 2023 Nov 29;18(11):e0294732. doi: 10.1371/journal.pone.0294732. eCollection 2023. PLoS One. 2023. PMID: 38019799 Free PMC article.
-
Single stranded DNA annealing is a conserved activity of telomere resolvases.PLoS One. 2021 Feb 4;16(2):e0246212. doi: 10.1371/journal.pone.0246212. eCollection 2021. PLoS One. 2021. PMID: 33539370 Free PMC article.
-
Design and characterization of hyperactive mutants of the Agrobacterium tumefaciens telomere resolvase, TelA.PLoS One. 2024 Jul 25;19(7):e0307590. doi: 10.1371/journal.pone.0307590. eCollection 2024. PLoS One. 2024. PMID: 39052566 Free PMC article.
-
Linear plasmids in Klebsiella and other Enterobacteriaceae.Microb Genom. 2022 Apr;8(4):000807. doi: 10.1099/mgen.0.000807. Microb Genom. 2022. PMID: 35416146 Free PMC article.
-
The Borrelia burgdorferi telomere resolvase, ResT, possesses ATP-dependent DNA unwinding activity.Nucleic Acids Res. 2017 Feb 17;45(3):1319-1329. doi: 10.1093/nar/gkw1243. Nucleic Acids Res. 2017. PMID: 28180323 Free PMC article.
References
-
- Ravin N.V., Strakhova T.S., Kuprianov V.V. The protelomerase of the phage-plasmid N15 is responsible for its maintenance in linear form. J. Mol. Biol. 2001;312:899–906. - PubMed
-
- Hertwig S., Klein I., Lurz R., Lanka E., Appel B. PY54, a linear plasmid prophage of Yersinia enterocolitica with covalently closed ends. Mol. Microbiol. 2003;48:989–1003. - PubMed
-
- Huang W.M., Joss L., Hsieh T., Casjens S. Protelomerase uses a topoisomerase IB/Y-recombinase type mechanism to generate DNA hairpin ends. J. Mol. Biol. 2004;337:77–92. - PubMed
-
- Picardeau M., Lobry J.R., Hinnebusch B.J. Physical mapping of an origin of bidirectional replication at the centre of the Borrelia burgdorferi linear chromosome. Mol. Microbiol. 1999;32:437–445. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

