Structural constraints and dynamics of bacterial cell wall architecture
- PMID: 26005443
- PMCID: PMC4424881
- DOI: 10.3389/fmicb.2015.00449
Structural constraints and dynamics of bacterial cell wall architecture
Abstract
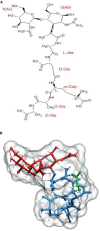
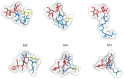
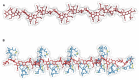
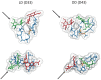
The peptidoglycan wall (PG) is a unique structure which confers physical strength and defined shape to bacteria. It consists of a net-like macromolecule of peptide interlinked glycan chains overlying the cell membrane. The structure and layout of the PG dictates that the wall has to be continuously modified as bacteria go through division, morphological differentiation, and adaptive responses. The PG is poorly known in structural terms. However, to understand morphogenesis a precise knowledge of glycan strand arrangement and of local effects of the different kinds of subunits is essential. The scarcity of data led to a conception of the PG as a regular, highly ordered structure which strongly influenced growth models. Here, we review the structure of the PG to define a more realistic conceptual framework. We discuss the consequences of the plasticity of murein architecture in morphogenesis and try to define a set of minimal structural constraints that must be fulfilled by any model to be compatible with present day information.
Keywords: HPLC; cell wall; chain length; cross-link; muropeptides; peptidoglycan; structure.
Figures
Similar articles
-
Peptidoglycan at its peaks: how chromatographic analyses can reveal bacterial cell wall structure and assembly.Mol Microbiol. 2013 Jul;89(1):1-13. doi: 10.1111/mmi.12266. Epub 2013 Jun 3. Mol Microbiol. 2013. PMID: 23679048 Free PMC article. Review.
-
Layered murein revisited: a fundamentally new concept of bacterial cell wall structure, biogenesis and function.Med Microbiol Immunol. 1999 Mar;187(3):173-81. doi: 10.1007/s004300050090. Med Microbiol Immunol. 1999. PMID: 10206149 Review.
-
Towards an automated analysis of bacterial peptidoglycan structure.Anal Bioanal Chem. 2017 Jan;409(2):551-560. doi: 10.1007/s00216-016-9857-5. Epub 2016 Aug 13. Anal Bioanal Chem. 2017. PMID: 27520322 Free PMC article.
-
Peptidoglycan hydrolase of an unusual cross-link cleavage specificity contributes to bacterial cell wall synthesis.Proc Natl Acad Sci U S A. 2019 Apr 16;116(16):7825-7830. doi: 10.1073/pnas.1816893116. Epub 2019 Apr 2. Proc Natl Acad Sci U S A. 2019. PMID: 30940749 Free PMC article.
-
Tertiary structure of Staphylococcus aureus cell wall murein.J Bacteriol. 2004 Nov;186(21):7141-8. doi: 10.1128/JB.186.21.7141-7148.2004. J Bacteriol. 2004. PMID: 15489425 Free PMC article.
Cited by
-
Interplay between Peptidoglycan Biology and Virulence in Gram-Negative Pathogens.Microbiol Mol Biol Rev. 2018 Sep 12;82(4):e00033-18. doi: 10.1128/MMBR.00033-18. Print 2018 Dec. Microbiol Mol Biol Rev. 2018. PMID: 30209071 Free PMC article. Review.
-
Modeling Novel Putative Drugs and Vaccine Candidates against Tick-Borne Pathogens: A Subtractive Proteomics Approach.Vet Sci. 2020 Sep 7;7(3):129. doi: 10.3390/vetsci7030129. Vet Sci. 2020. PMID: 32906620 Free PMC article.
-
Peptidoglycan precursor synthesis along the sidewall of pole-growing mycobacteria.Elife. 2018 Sep 10;7:e37243. doi: 10.7554/eLife.37243. Elife. 2018. PMID: 30198841 Free PMC article.
-
Mechanisms of substrate recognition by a typhoid toxin secretion-associated muramidase.Elife. 2020 Jan 20;9:e53473. doi: 10.7554/eLife.53473. Elife. 2020. PMID: 31958059 Free PMC article.
-
Purification and HPLC Analysis of Cell Wall Muropeptides from Caulobacter crescentus.Bio Protoc. 2019 Nov 5;9(21):e3421. doi: 10.21769/BioProtoc.3421. eCollection 2019 Nov 5. Bio Protoc. 2019. PMID: 33654919 Free PMC article.
References
-
- Barnikel G., Naumann D., Bradaczek H., Labischinski H., Giesbrecht P. (1983). “Computer-aided molecular modelling of the three dimensional structure of bacterial peptidoglycan,” in The Target of Penicillin: The Murein Sacculus of Bacterial Cell Walls, Architecture and Growth, edsHakenbeck J.-V. H. R., Labischinski H. (Berlin: de Gruyter; ), 61–66.

