Bayesian mixture analysis for metagenomic community profiling
- PMID: 26002885
- PMCID: PMC4565032
- DOI: 10.1093/bioinformatics/btv317
Bayesian mixture analysis for metagenomic community profiling
Abstract
Motivation: Deep sequencing of clinical samples is now an established tool for the detection of infectious pathogens, with direct medical applications. The large amount of data generated produces an opportunity to detect species even at very low levels, provided that computational tools can effectively profile the relevant metagenomic communities. Data interpretation is complicated by the fact that short sequencing reads can match multiple organisms and by the lack of completeness of existing databases, in particular for viral pathogens. Here we present metaMix, a Bayesian mixture model framework for resolving complex metagenomic mixtures. We show that the use of parallel Monte Carlo Markov chains for the exploration of the species space enables the identification of the set of species most likely to contribute to the mixture.
Results: We demonstrate the greater accuracy of metaMix compared with relevant methods, particularly for profiling complex communities consisting of several related species. We designed metaMix specifically for the analysis of deep transcriptome sequencing datasets, with a focus on viral pathogen detection; however, the principles are generally applicable to all types of metagenomic mixtures.
Availability and implementation: metaMix is implemented as a user friendly R package, freely available on CRAN: http://cran.r-project.org/web/packages/metaMix
Contact: sofia.morfopoulou.10@ucl.ac.uk
Supplementary information: Supplementary data are available at Bionformatics online.
© The Author 2015. Published by Oxford University Press.
Figures
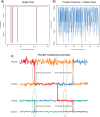
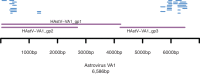
Similar articles
-
Sigma: strain-level inference of genomes from metagenomic analysis for biosurveillance.Bioinformatics. 2015 Jan 15;31(2):170-7. doi: 10.1093/bioinformatics/btu641. Epub 2014 Sep 29. Bioinformatics. 2015. PMID: 25266224 Free PMC article.
-
PAIPline: pathogen identification in metagenomic and clinical next generation sequencing samples.Bioinformatics. 2018 Sep 1;34(17):i715-i721. doi: 10.1093/bioinformatics/bty595. Bioinformatics. 2018. PMID: 30423069 Free PMC article.
-
ViromeScan: a new tool for metagenomic viral community profiling.BMC Genomics. 2016 Mar 1;17:165. doi: 10.1186/s12864-016-2446-3. BMC Genomics. 2016. PMID: 26932765 Free PMC article.
-
From genomics to metagenomics.Curr Opin Biotechnol. 2012 Feb;23(1):72-6. doi: 10.1016/j.copbio.2011.12.017. Epub 2012 Jan 5. Curr Opin Biotechnol. 2012. PMID: 22227326 Review.
-
Web Resources for Metagenomics Studies.Genomics Proteomics Bioinformatics. 2015 Oct;13(5):296-303. doi: 10.1016/j.gpb.2015.10.003. Epub 2015 Nov 18. Genomics Proteomics Bioinformatics. 2015. PMID: 26602607 Free PMC article. Review.
Cited by
-
Report of the third conference on next-generation sequencing for adventitious virus detection in biologics for humans and animals.Biologicals. 2023 Aug;83:101696. doi: 10.1016/j.biologicals.2023.101696. Epub 2023 Jul 19. Biologicals. 2023. PMID: 37478506 Free PMC article.
-
Metataxonomic and Metagenomic Approaches vs. Culture-Based Techniques for Clinical Pathology.Front Microbiol. 2016 Apr 7;7:484. doi: 10.3389/fmicb.2016.00484. eCollection 2016. Front Microbiol. 2016. PMID: 27092134 Free PMC article.
-
Metagenomic analysis of a blood stain from the French revolutionary Jean-Paul Marat (1743-1793).Infect Genet Evol. 2020 Jun;80:104209. doi: 10.1016/j.meegid.2020.104209. Epub 2020 Jan 29. Infect Genet Evol. 2020. PMID: 32004756 Free PMC article.
-
Metagenomic evidence for a polymicrobial signature of sepsis.Microb Genom. 2021 Sep;7(9):000642. doi: 10.1099/mgen.0.000642. Microb Genom. 2021. PMID: 34477543 Free PMC article.
-
Evaluating metagenomics and targeted approaches for diagnosis and surveillance of viruses.Genome Med. 2024 Sep 9;16(1):111. doi: 10.1186/s13073-024-01380-x. Genome Med. 2024. PMID: 39252069 Free PMC article.
References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

