XPD localizes in mitochondria and protects the mitochondrial genome from oxidative DNA damage
- PMID: 25969448
- PMCID: PMC4477675
- DOI: 10.1093/nar/gkv472
XPD localizes in mitochondria and protects the mitochondrial genome from oxidative DNA damage
Abstract
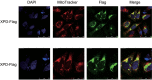
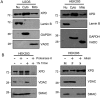
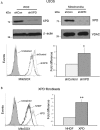
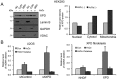
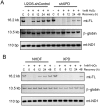
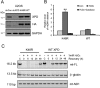
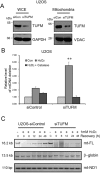
Xeroderma pigmentosum group D (XPD/ERCC2) encodes an ATP-dependent helicase that plays essential roles in both transcription and nucleotide excision repair of nuclear DNA, however, whether or not XPD exerts similar functions in mitochondria remains elusive. In this study, we provide the first evidence that XPD is localized in the inner membrane of mitochondria, and cells under oxidative stress showed an enhanced recruitment of XPD into mitochondrial compartment. Furthermore, mitochondrial reactive oxygen species production and levels of oxidative stress-induced mitochondrial DNA (mtDNA) common deletion were significantly elevated, whereas capacity for oxidative damage repair of mtDNA was markedly reduced in both XPD-suppressed human osteosarcoma (U2OS) cells and XPD-deficient human fibroblasts. Immunoprecipitation-mass spectrometry analysis was used to identify interacting factor(s) with XPD and TUFM, a mitochondrial Tu translation elongation factor was detected to be physically interacted with XPD. Similar to the findings in XPD-deficient cells, mitochondrial common deletion and oxidative damage repair capacity in U2OS cells were found to be significantly altered after TUFM knock-down. Our findings clearly demonstrate that XPD plays crucial role(s) in protecting mitochondrial genome stability by facilitating an efficient repair of oxidative DNA damage in mitochondria.
© The Author(s) 2015. Published by Oxford University Press on behalf of Nucleic Acids Research.
Figures

Similar articles
-
Role of Xeroderma pigmentosum D (XPD) protein in genome maintenance in human cells under oxidative stress.Mutat Res Genet Toxicol Environ Mutagen. 2022 Apr-May;876-877:503444. doi: 10.1016/j.mrgentox.2022.503444. Epub 2022 Jan 15. Mutat Res Genet Toxicol Environ Mutagen. 2022. PMID: 35483790
-
MMS19 localizes to mitochondria and protects the mitochondrial genome from oxidative damage.Biochem Cell Biol. 2018 Feb;96(1):44-49. doi: 10.1139/bcb-2017-0149. Epub 2017 Oct 16. Biochem Cell Biol. 2018. PMID: 29035693
-
Increased Oxidative Damage and Reduced DNA Repair Enzyme XPD Involvement in High Glucose-Mediated Enhancement of Levobupivacaine-Induced Neurotoxicity.Neurochem Res. 2015 Sep;40(9):1919-28. doi: 10.1007/s11064-015-1685-z. Epub 2015 Aug 12. Neurochem Res. 2015. PMID: 26264262
-
The XPD helicase: XPanDing archaeal XPD structures to get a grip on human DNA repair.Biol Chem. 2010 Jul;391(7):761-5. doi: 10.1515/BC.2010.076. Biol Chem. 2010. PMID: 20482310 Review.
-
Mitochondria-nucleus network for genome stability.Free Radic Biol Med. 2015 May;82:73-104. doi: 10.1016/j.freeradbiomed.2015.01.013. Epub 2015 Jan 30. Free Radic Biol Med. 2015. PMID: 25640729 Review.
Cited by
-
Mitochondrial DNA Instability in Mammalian Cells.Antioxid Redox Signal. 2022 May;36(13-15):885-905. doi: 10.1089/ars.2021.0091. Epub 2021 Jul 2. Antioxid Redox Signal. 2022. PMID: 34015960 Free PMC article. Review.
-
Xeroderma Pigmentosum Group D (XPD) Inhibits the Proliferation Cycle of Vascular Smooth Muscle Cell (VSMC) by Activating Glycogen Synthase Kinase 3β (GSK3β).Med Sci Monit. 2018 Aug 27;24:5951-5959. doi: 10.12659/MSM.909614. Med Sci Monit. 2018. PMID: 30146633 Free PMC article.
-
DNA polymerase β: A missing link of the base excision repair machinery in mammalian mitochondria.DNA Repair (Amst). 2017 Dec;60:77-88. doi: 10.1016/j.dnarep.2017.10.011. Epub 2017 Oct 28. DNA Repair (Amst). 2017. PMID: 29100041 Free PMC article.
-
Dynamic features of human mitochondrial DNA maintenance and transcription.Front Cell Dev Biol. 2022 Sep 7;10:984245. doi: 10.3389/fcell.2022.984245. eCollection 2022. Front Cell Dev Biol. 2022. PMID: 36158192 Free PMC article. Review.
-
The RXFP3 receptor is functionally associated with cellular responses to oxidative stress and DNA damage.Aging (Albany NY). 2019 Dec 3;11(23):11268-11313. doi: 10.18632/aging.102528. Epub 2019 Dec 3. Aging (Albany NY). 2019. PMID: 31794429 Free PMC article.
References
-
- Sung P., Bailly V., Weber C., Thompson L.H., Prakash L., Prakash S. Human xeroderma pigmentosum group D gene encodes a DNA helicase. Nature. 1993;365:852–855. - PubMed
-
- Zurita M., Merino C. The transcriptional complexity of the TFIIH complex. Trends Genet. 2003;19:578–584. - PubMed
-
- Tirode F., Busso D., Coin F., Egly J.M. Reconstitution of the transcription factor TFIIH: assignment of functions for the three enzymatic subunits, XPB, XPD, and cdk7. Mol. Cell. 1999;3:87–95. - PubMed
-
- Broughton B.C., Thompson A.F., Harcourt S.A., Vermeulen W., Hoeijmakers J.H.J., Botta E., Stefanini M., King M.D., Weber C.A., Cole J., et al. Molecular and cellular analysis of the DNA-repair defect in a patient in xeroderma-pigmentosum complementation group-D who has the clinical-features of xeroderma-pigmentosum and Cockayne-syndrome. Am. J. Hum. Genet. 1995;56:167–174. - PMC - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials

