Estrogen receptor beta impacts hormone-induced alternative mRNA splicing in breast cancer cells
- PMID: 25956916
- PMCID: PMC4424892
- DOI: 10.1186/s12864-015-1541-1
Estrogen receptor beta impacts hormone-induced alternative mRNA splicing in breast cancer cells
Abstract
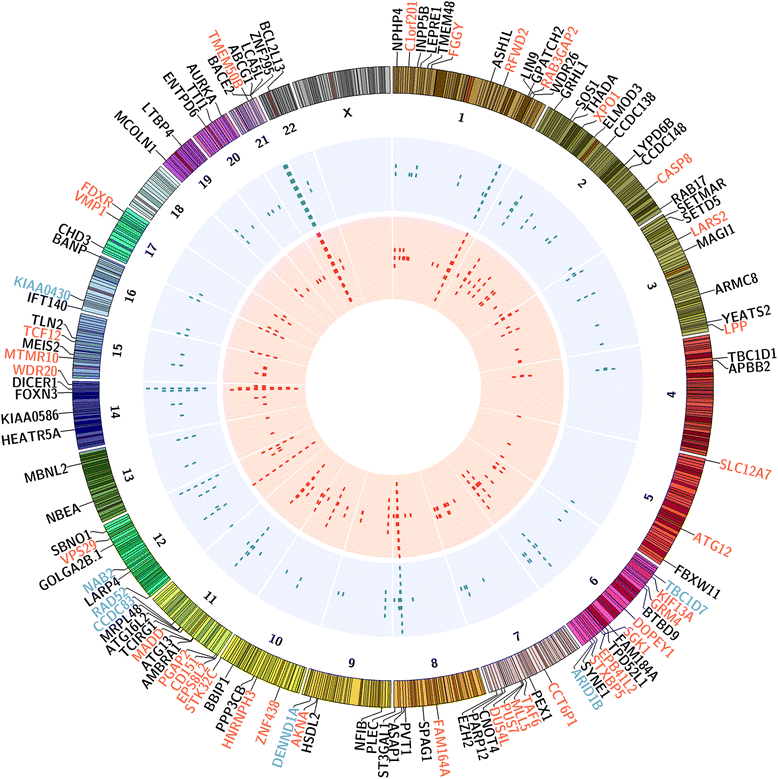
Background: Estrogens play an important role in breast cancer (BC) development and progression; when the two isoforms of the estrogen receptor (ERα and ERβ) are co-expressed each of them mediate specific effects of these hormones in BC cells. ERβ has been suggested to exert an antagonist role toward the oncogenic activities of ERα, and for this reason it is considered an oncosuppressor. As clinical evidence regarding a prognostic role for this receptor subtype in hormone-responsive BC is still limited and conflicting, more knowledge is required on the biological functions of ERβ in cancer cells. We have previously described the ERβ and ERα interactomes from BC cells, identifying specific and distinct patterns of protein interactions for the two receptors. In particular, we identified factors involved in mRNA splicing and maturation as important components of both ERα and ERβ pathways. Guided by these findings, here we performed RNA sequencing to investigate in depth the differences in the early transcriptional events and RNA splicing patterns induced by estradiol in cells expressing ERα alone or ERα and ERβ.
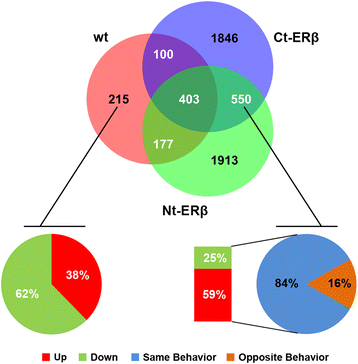
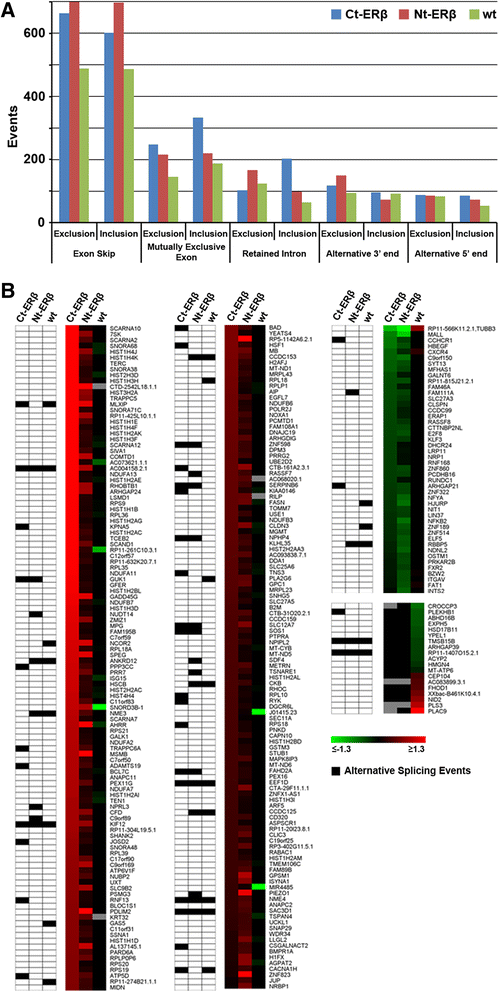
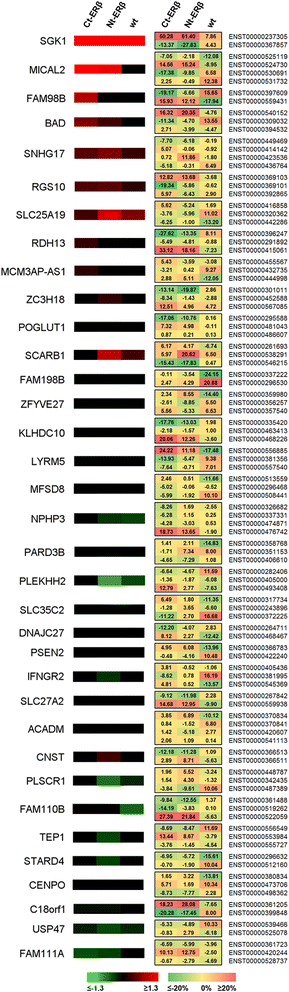
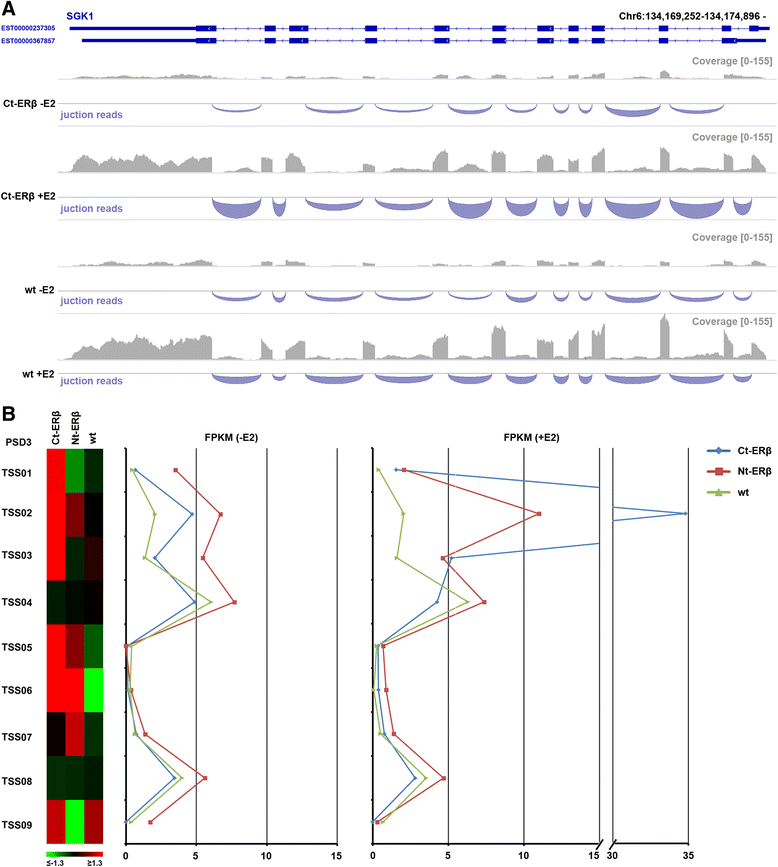
Results: Exon skipping was the most abundant splicing event in the post-transcriptional regulation by estradiol. We identified several splicing events induced by ERα alone and by ERα+ERβ, demonstrating for the first time that ERβ significantly affects estrogen-induced splicing in BC cells, as revealed by modification of a subset of ERα-dependent splicing by ERβ, as well as by the presence of splicing isoforms only in ERβ+cells. In particular, we observed that ERβ+BC cell lines exhibited around 2-fold more splicing events than the ERβ- cells. Interestingly, we identified putative direct targets of ERβ-mediated alternative splicing by correlating the genomic locations of ERβ and ERα binding sites with estradiol-induced differential splicing in the corresponding genes.
Conclusions: Taken together, these results demonstrate that ERβ significantly affects estrogen-induced early transcription and mRNA splicing in hormone-responsive BC cells, providing novel information on the biological role of ERβ in these tumors.
Figures
Similar articles
-
Transcriptional regulation of vascular endothelial growth factor by estradiol and tamoxifen in breast cancer cells: a complex interplay between estrogen receptors alpha and beta.Cancer Res. 2002 Sep 1;62(17):4977-84. Cancer Res. 2002. PMID: 12208749
-
Direct regulation of microRNA biogenesis and expression by estrogen receptor beta in hormone-responsive breast cancer.Oncogene. 2012 Sep 20;31(38):4196-206. doi: 10.1038/onc.2011.583. Epub 2012 Jan 9. Oncogene. 2012. PMID: 22231442
-
Indole-3-carbinol selectively uncouples expression and activity of estrogen receptor subtypes in human breast cancer cells.Mol Endocrinol. 2006 Dec;20(12):3070-82. doi: 10.1210/me.2005-0263. Epub 2006 Aug 10. Mol Endocrinol. 2006. PMID: 16901971
-
Estrogen receptor alpha negative breast cancer patients: estrogen receptor beta as a therapeutic target.J Steroid Biochem Mol Biol. 2008 Mar;109(1-2):1-10. doi: 10.1016/j.jsbmb.2007.12.010. Epub 2007 Dec 8. J Steroid Biochem Mol Biol. 2008. PMID: 18243688 Review.
-
Regulation of specific target genes and biological responses by estrogen receptor subtype agonists.Curr Opin Pharmacol. 2010 Dec;10(6):629-36. doi: 10.1016/j.coph.2010.09.009. Epub 2010 Oct 14. Curr Opin Pharmacol. 2010. PMID: 20951642 Free PMC article. Review.
Cited by
-
Epigenetic alterations of CYP19A1 gene in Cumulus cells and its relevance to infertility in endometriosis.J Assist Reprod Genet. 2016 Aug;33(8):1105-13. doi: 10.1007/s10815-016-0727-z. Epub 2016 May 11. J Assist Reprod Genet. 2016. PMID: 27167072 Free PMC article.
-
Regulating the regulator: a survey of mechanisms from transcription to translation controlling expression of mammalian cell cycle kinase Aurora A.Open Biol. 2022 Sep;12(9):220134. doi: 10.1098/rsob.220134. Epub 2022 Sep 7. Open Biol. 2022. PMID: 36067794 Free PMC article. Review.
-
Splicing factors control triple-negative breast cancer cell mitosis through SUN2 interaction and sororin intron retention.J Exp Clin Cancer Res. 2021 Mar 1;40(1):82. doi: 10.1186/s13046-021-01863-4. J Exp Clin Cancer Res. 2021. PMID: 33648524 Free PMC article.
-
Estrogen Receptors in Epithelial-Mesenchymal Transition of Prostate Cancer.Cancers (Basel). 2019 Sep 23;11(10):1418. doi: 10.3390/cancers11101418. Cancers (Basel). 2019. PMID: 31548498 Free PMC article. Review.
-
Estrogen receptor β exerts tumor suppressive effects in prostate cancer through repression of androgen receptor activity.PLoS One. 2020 May 15;15(5):e0226057. doi: 10.1371/journal.pone.0226057. eCollection 2020. PLoS One. 2020. PMID: 32413024 Free PMC article.
References
-
- Ferlay J, Soerjomataram I, Dikshit R, Eser S, Mathers C, Rebelo M, et al. Cancer incidence and mortality worldwide: sources, methods and major patterns in GLOBOCAN 2012. Int J Cancer. 2015;136(5):E359-86. - PubMed
-
- Nilsson S, Makela S, Treuter E, Tujague M, Thomsen J, Andersson G, et al. Mechanisms of estrogen action. Physiol Rev. 2001;81(4):1535–1565. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases
Research Materials

