Three-dimensional, soft neotissue arrays as high throughput platforms for the interrogation of engineered tissue environments
- PMID: 25956850
- PMCID: PMC4444363
- DOI: 10.1016/j.biomaterials.2015.04.036
Three-dimensional, soft neotissue arrays as high throughput platforms for the interrogation of engineered tissue environments
Erratum in
- Biomaterials. 2015 Oct;67:204
Abstract
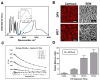
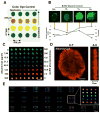
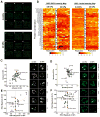
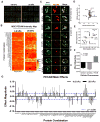
Local signals from tissue-specific extracellular matrix (ECM) microenvironments, including matrix adhesive ligand, mechanical elasticity and micro-scale geometry, are known to instruct a variety of stem cell differentiation processes. Likewise, these signals converge to provide multifaceted, mechanochemical cues for highly-specific tissue morphogenesis or regeneration. Despite accumulated knowledge about the individual and combined roles of various mechanochemical ECM signals in stem cell activities on 2-dimensional matrices, the understandings of morphogenetic or regenerative 3-dimenstional tissue microenvironments remain very limited. To that end, we established high-throughput platforms based on soft, fibrous matrices with various combinatorial ECM proteins meanwhile highly-tunable in elasticity and 3-dimensional geometry. To demonstrate the utility of our platform, we evaluated 64 unique combinations of 6 ECM proteins (collagen I, collagen III, collagen IV, laminin, fibronectin, and elastin) on the adhesion, spreading and fate commitment of mesenchymal stem cell (MSCs) under two substrate stiffness (4.6 kPa, 20 kPa). Using this technique, we identified several neotissue microenvironments supporting MSC adhesion, spreading and differentiation toward early vascular lineages. Manipulation of the matrix properties, such as elasticity and geometry, in concert with ECM proteins will permit the investigation of multiple and distinct MSC environments. This paper demonstrates the practical application of high through-put technology to facilitate the screening of a variety of engineered microenvironments with the aim to instruct stem cell differentiation.
Keywords: 3-Dimensional cell culture; Extracellular matrix; High-throughput screening; Stem cell differentiation.
Copyright © 2015 Elsevier Ltd. All rights reserved.
Figures
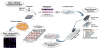
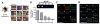

Similar articles
-
XanoMatrix surfaces as scaffolds for mesenchymal stem cell culture and growth.Int J Nanomedicine. 2016 Jun 7;11:2655-61. doi: 10.2147/IJN.S101838. eCollection 2016. Int J Nanomedicine. 2016. PMID: 27354795 Free PMC article.
-
Matrix directed adipogenesis and neurogenesis of mesenchymal stem cells derived from adipose tissue and bone marrow.Acta Biomater. 2016 Sep 15;42:46-55. doi: 10.1016/j.actbio.2016.06.037. Epub 2016 Jun 29. Acta Biomater. 2016. PMID: 27375285 Free PMC article.
-
Extracellular matrix microarrays to study inductive signaling for endoderm specification.Acta Biomater. 2016 Apr 1;34:30-40. doi: 10.1016/j.actbio.2016.02.014. Epub 2016 Feb 12. Acta Biomater. 2016. PMID: 26883775 Free PMC article.
-
Union is strength: matrix elasticity and microenvironmental factors codetermine stem cell differentiation fate.Cell Tissue Res. 2015 Sep;361(3):657-68. doi: 10.1007/s00441-015-2190-z. Epub 2015 May 9. Cell Tissue Res. 2015. PMID: 25956590 Review.
-
A New Chapter for Mesenchymal Stem Cells: Decellularized Extracellular Matrices.Stem Cell Rev Rep. 2017 Oct;13(5):587-597. doi: 10.1007/s12015-017-9757-x. Stem Cell Rev Rep. 2017. PMID: 28766102 Review.
Cited by
-
Biomimetic soft fibrous hydrogels for contractile and pharmacologically responsive smooth muscle.Acta Biomater. 2018 Jul 1;74:121-130. doi: 10.1016/j.actbio.2018.05.015. Epub 2018 May 16. Acta Biomater. 2018. PMID: 29753912 Free PMC article.
-
Convergence of Highly Resolved and Rapid Screening Platforms with Dynamically Engineered, Cell Phenotype-Prescriptive Biomaterials.Curr Pharmacol Rep. 2016 Jun;2(3):142-151. doi: 10.1007/s40495-016-0057-y. Epub 2016 Mar 18. Curr Pharmacol Rep. 2016. PMID: 27482508 Free PMC article.
-
Combinatorial hydrogels with biochemical gradients for screening 3D cellular microenvironments.Nat Commun. 2018 Feb 9;9(1):614. doi: 10.1038/s41467-018-03021-5. Nat Commun. 2018. PMID: 29426836 Free PMC article.
-
Functional and Biomimetic Materials for Engineering of the Three-Dimensional Cell Microenvironment.Chem Rev. 2017 Oct 25;117(20):12764-12850. doi: 10.1021/acs.chemrev.7b00094. Epub 2017 Oct 9. Chem Rev. 2017. PMID: 28991456 Free PMC article. Review.
-
Pulmonary Arterial Stiffness: Toward a New Paradigm in Pulmonary Arterial Hypertension Pathophysiology and Assessment.Curr Hypertens Rep. 2016 Jan;18(1):4. doi: 10.1007/s11906-015-0609-2. Curr Hypertens Rep. 2016. PMID: 26733189
References
-
- Reilly GC, Engler AJ. Intrinsic extracellular matrix properties regulate stem cell differentiation. Biomechanics. 2010;43:55–62. - PubMed
-
- da Silva Meirelles L, Caplan AI, Nardi NB. In search of the in vivo identity of mesenchymal stem cells. Stem Cells. 2008;26:2287. - PubMed
-
- Suzuki S, Narita Y, Yamawaki A, Murase Y, Satake M, Mutsuga M, et al. Effects of extracellular matrix on differentiation of human bone marrow-derived mesenchymal stem cells into smooth muscle cell lineage: utility for cardiovascular tissue engineering. Cells Tissues Organs. 2010;191(4):269–280. - PubMed
-
- Engler JA, Sen S, Sweeney HL, Discher DE. Matrix Elasticity Directs Stem Cell Lineage Specification. Cell. 2006;126:677–689. - PubMed
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

