An integrative framework for the identification of double minute chromosomes using next generation sequencing data
- PMID: 25953282
- PMCID: PMC4423570
- DOI: 10.1186/1471-2156-16-S2-S1
An integrative framework for the identification of double minute chromosomes using next generation sequencing data
Abstract
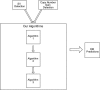
Background: Double minute chromosomes are circular fragments of DNA whose presence is associated with the onset of certain cancers. Double minutes are lethal, as they are highly amplified and typically contain oncogenes. Locating double minutes can supplement the process of cancer diagnosis, and it can help to identify therapeutic targets. However, there is currently a dearth of computational methods available to identify double minutes. We propose a computational framework for the idenfication of double minute chromosomes using next-generation sequencing data. Our framework integrates predictions from algorithms that detect DNA copy number variants, and it also integrates predictions from algorithms that locate genomic structural variants. This information is used by a graph-based algorithm to predict the presence of double minute chromosomes.
Results: Using a previously published copy number variant algorithm and two structural variation prediction algorithms, we implemented our framework and tested it on a dataset consisting of simulated double minute chromosomes. Our approach uncovered double minutes with high accuracy, demonstrating its plausibility.
Conclusions: Although we only tested the framework with three programs (RDXplorer, BreakDancer, Delly), it can be extended to incorporate results from programs that 1) detect amplified copy number and from programs that 2) detect genomic structural variants like deletions, translocations, inversions, and tandem repeats. The software that implements the framework can be accessed here: https://github.com/mhayes20/DMFinder
Figures







Similar articles
-
HolistIC: leveraging Hi-C and whole genome shotgun sequencing for double minute chromosome discovery.Bioinformatics. 2022 Feb 7;38(5):1208-1215. doi: 10.1093/bioinformatics/btab816. Bioinformatics. 2022. PMID: 34888626 Free PMC article.
-
RAPTR-SV: a hybrid method for the detection of structural variants.Bioinformatics. 2015 Jul 1;31(13):2084-90. doi: 10.1093/bioinformatics/btv086. Epub 2015 Feb 16. Bioinformatics. 2015. PMID: 25686638
-
Detecting large deletions at base pair level by combining split read and paired read data.BMC Bioinformatics. 2017 Oct 16;18(Suppl 12):413. doi: 10.1186/s12859-017-1829-z. BMC Bioinformatics. 2017. PMID: 29072143 Free PMC article.
-
Inferring the global structure of chromosomes from structural variations.BMC Genomics. 2015;16 Suppl 2(Suppl 2):S13. doi: 10.1186/1471-2164-16-S2-S13. Epub 2015 Jan 21. BMC Genomics. 2015. PMID: 25707904 Free PMC article.
-
Life of double minutes: generation, maintenance, and elimination.Chromosoma. 2022 Sep;131(3):107-125. doi: 10.1007/s00412-022-00773-4. Epub 2022 Apr 30. Chromosoma. 2022. PMID: 35487993 Free PMC article. Review.
Cited by
-
HolistIC: leveraging Hi-C and whole genome shotgun sequencing for double minute chromosome discovery.Bioinformatics. 2022 Feb 7;38(5):1208-1215. doi: 10.1093/bioinformatics/btab816. Bioinformatics. 2022. PMID: 34888626 Free PMC article.
-
Structure and evolution of double minutes in diagnosis and relapse brain tumors.Acta Neuropathol. 2019 Jan;137(1):123-137. doi: 10.1007/s00401-018-1912-1. Epub 2018 Sep 28. Acta Neuropathol. 2019. PMID: 30267146 Free PMC article.
-
Molecular structure and evolution mechanism of two populations of double minutes in human colorectal cancer cells.J Cell Mol Med. 2020 Dec;24(24):14205-14216. doi: 10.1111/jcmm.16035. Epub 2020 Oct 30. J Cell Mol Med. 2020. PMID: 33124133 Free PMC article.
-
Bioinformatics advances in eccDNA identification and analysis.Oncogene. 2024 Oct;43(41):3021-3036. doi: 10.1038/s41388-024-03138-6. Epub 2024 Aug 29. Oncogene. 2024. PMID: 39209966 Review.
-
Overexpression of hsa-miR-186 induces chromosomal instability in arsenic-exposed human keratinocytes.Toxicol Appl Pharmacol. 2019 Sep 1;378:114614. doi: 10.1016/j.taap.2019.114614. Epub 2019 Jun 6. Toxicol Appl Pharmacol. 2019. PMID: 31176655 Free PMC article.
References
-
- Storlazzi CT, Lonoce A, Guastadisegni MC, Trombetta D, D'Addabbo P, Daniele G, L'Abbate A, Macchia G, Surace C, Kok K, Ullmann R, Purgato S, Palumbo O, Carella M, Ambros PF, Rocchi M. Gene amplification as double minutes or homogeneously staining regions in solid tumors: origin and structure. Genome Res. 2010;20(9):1198–1206. doi: 10.1101/gr.106252.110. - DOI - PMC - PubMed
MeSH terms
LinkOut - more resources
Full Text Sources
Other Literature Sources

