Oncogenic and RASopathy-associated K-RAS mutations relieve membrane-dependent occlusion of the effector-binding site
- PMID: 25941399
- PMCID: PMC4450377
- DOI: 10.1073/pnas.1419895112
Oncogenic and RASopathy-associated K-RAS mutations relieve membrane-dependent occlusion of the effector-binding site
Abstract
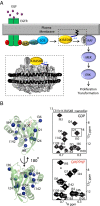
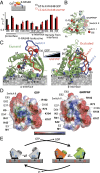
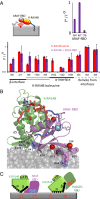
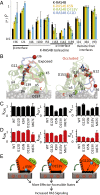
K-RAS4B (Kirsten rat sarcoma viral oncogene homolog 4B) is a prenylated, membrane-associated GTPase protein that is a critical switch for the propagation of growth factor signaling pathways to diverse effector proteins, including rapidly accelerated fibrosarcoma (RAF) kinases and RAS-related protein guanine nucleotide dissociation stimulator (RALGDS) proteins. Gain-of-function KRAS mutations occur frequently in human cancers and predict poor clinical outcome, whereas germ-line mutations are associated with developmental syndromes. However, it is not known how these mutations affect K-RAS association with biological membranes or whether this impacts signal transduction. Here, we used solution NMR studies of K-RAS4B tethered to nanodiscs to investigate lipid bilayer-anchored K-RAS4B and its interactions with effector protein RAS-binding domains (RBDs). Unexpectedly, we found that the effector-binding region of activated K-RAS4B is occluded by interaction with the membrane in one of the NMR-observable, and thus highly populated, conformational states. Binding of the RAF isoform ARAF and RALGDS RBDs induced marked reorientation of K-RAS4B from the occluded state to RBD-specific effector-bound states. Importantly, we found that two Noonan syndrome-associated mutations, K5N and D153V, which do not affect the GTPase cycle, relieve the occluded orientation by directly altering the electrostatics of two membrane interaction surfaces. Similarly, the most frequent KRAS oncogenic mutation G12D also drives K-RAS4B toward an exposed configuration. Further, the D153V and G12D mutations increase the rate of association of ARAF-RBD with lipid bilayer-tethered K-RAS4B. We revealed a mechanism of K-RAS4B autoinhibition by membrane sequestration of its effector-binding site, which can be disrupted by disease-associated mutations. Stabilizing the autoinhibitory interactions between K-RAS4B and the membrane could be an attractive target for anticancer drug discovery.
Keywords: KRAS; Noonan syndrome; lipid bilayer nanodisc; nuclear magnetic resonance; oncogenic mutation.
Conflict of interest statement
The authors declare no conflict of interest.
Figures
Similar articles
-
GTP Binding and Oncogenic Mutations May Attenuate Hypervariable Region (HVR)-Catalytic Domain Interactions in Small GTPase K-Ras4B, Exposing the Effector Binding Site.J Biol Chem. 2015 Nov 27;290(48):28887-900. doi: 10.1074/jbc.M115.664755. Epub 2015 Oct 9. J Biol Chem. 2015. PMID: 26453300 Free PMC article.
-
Oncogenic K-Ras4B Dimerization Enhances Downstream Mitogen-activated Protein Kinase Signaling.J Mol Biol. 2020 Feb 14;432(4):1199-1215. doi: 10.1016/j.jmb.2020.01.002. Epub 2020 Jan 10. J Mol Biol. 2020. PMID: 31931009 Free PMC article.
-
Inhibition of K-RAS4B by a Unique Mechanism of Action: Stabilizing Membrane-Dependent Occlusion of the Effector-Binding Site.Cell Chem Biol. 2018 Nov 15;25(11):1327-1336.e4. doi: 10.1016/j.chembiol.2018.07.009. Epub 2018 Aug 16. Cell Chem Biol. 2018. PMID: 30122370
-
The Hypervariable Region of K-Ras4B Governs Molecular Recognition and Function.Int J Mol Sci. 2019 Nov 14;20(22):5718. doi: 10.3390/ijms20225718. Int J Mol Sci. 2019. PMID: 31739603 Free PMC article. Review.
-
Principles of K-Ras effector organization and the role of oncogenic K-Ras in cancer initiation through G1 cell cycle deregulation.Expert Rev Proteomics. 2015;12(6):669-82. doi: 10.1586/14789450.2015.1100079. Epub 2015 Oct 23. Expert Rev Proteomics. 2015. PMID: 26496174 Review.
Cited by
-
The hanging heart: How KRAS lures its prey to the membrane.Proc Natl Acad Sci U S A. 2020 Oct 27;117(43):26556-26558. doi: 10.1073/pnas.2017730117. Epub 2020 Sep 30. Proc Natl Acad Sci U S A. 2020. PMID: 32999064 Free PMC article. No abstract available.
-
Dynamically encoded reactivity of Ras enzymes: opening new frontiers for drug discovery.Cancer Metastasis Rev. 2020 Dec;39(4):1075-1089. doi: 10.1007/s10555-020-09917-3. Epub 2020 Aug 20. Cancer Metastasis Rev. 2020. PMID: 32815102 Free PMC article. Review.
-
Multivalent assembly of KRAS with the RAS-binding and cysteine-rich domains of CRAF on the membrane.Proc Natl Acad Sci U S A. 2020 Jun 2;117(22):12101-12108. doi: 10.1073/pnas.1914076117. Epub 2020 May 15. Proc Natl Acad Sci U S A. 2020. PMID: 32414921 Free PMC article.
-
Real-time monitoring of the reaction of KRAS G12C mutant specific covalent inhibitor by in vitro and in-cell NMR spectroscopy.Sci Rep. 2023 Nov 7;13(1):19253. doi: 10.1038/s41598-023-46623-w. Sci Rep. 2023. PMID: 37935773 Free PMC article.
-
New weapons to penetrate the armor: Novel reagents and assays developed at the NCI RAS Initiative to enable discovery of RAS therapeutics.Semin Cancer Biol. 2019 Feb;54:174-182. doi: 10.1016/j.semcancer.2018.02.006. Epub 2018 Feb 9. Semin Cancer Biol. 2019. PMID: 29432816 Free PMC article. Review.
References
-
- Tian T, et al. Plasma membrane nanoswitches generate high-fidelity Ras signal transduction. Nat Cell Biol. 2007;9(8):905–914. - PubMed
-
- Kapoor S, et al. The role of G-domain orientation and nucleotide state on the Ras isoform-specific membrane interaction. Eur Biophys J. 2012;41(10):801–813. - PubMed
-
- Weise K, et al. Membrane-mediated induction and sorting of K-Ras microdomain signaling platforms. J Am Chem Soc. 2011;133(4):880–887. - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
- Actions
- Actions
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials
Miscellaneous

