Emerging structural insights into the function of ionotropic glutamate receptors
- PMID: 25941168
- PMCID: PMC4464829
- DOI: 10.1016/j.tibs.2015.04.002
Emerging structural insights into the function of ionotropic glutamate receptors
Abstract
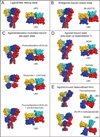
Ionotropic glutamate receptors (iGluRs) are ligand-gated ion channels that mediate excitatory neurotransmission crucial for brain development and function, including learning and memory formation. Recently a wealth of structural studies on iGluRs including AMPA receptors (AMPARs), kainate receptors, and NMDA receptors (NMDARs) became available. These studies showed structures of non-NMDARs including AMPAR and kainate receptor in various functional states, thereby providing the first visual sense of how non-NMDAR iGluRs may function in the context of homotetramers. Furthermore, they provided the first view of heterotetrameric NMDAR ion channels, and this illuminated the similarities with and differences from non-NMDARs, thus raising a mechanistic distinction between the two groups of iGluRs. We review mechanistic insights into iGluR functions gained through structural studies of multiple groups.
Keywords: ionotropic glutamate receptors; pharmacology; structural biology.
Copyright © 2015 Elsevier Ltd. All rights reserved.
Figures




Similar articles
-
Positron Emission Tomography (PET) Ligand Development for Ionotropic Glutamate Receptors: Challenges and Opportunities for Radiotracer Targeting N-Methyl-d-aspartate (NMDA), α-Amino-3-hydroxy-5-methyl-4-isoxazolepropionic Acid (AMPA), and Kainate Receptors.J Med Chem. 2019 Jan 24;62(2):403-419. doi: 10.1021/acs.jmedchem.8b00714. Epub 2018 Aug 27. J Med Chem. 2019. PMID: 30110164 Free PMC article. Review.
-
Chemoenzymatic synthesis of new 2,4-syn-functionalized (S)-glutamate analogues and structure-activity relationship studies at ionotropic glutamate receptors and excitatory amino acid transporters.J Med Chem. 2013 Feb 28;56(4):1614-28. doi: 10.1021/jm301433m. Epub 2013 Feb 15. J Med Chem. 2013. PMID: 23414088
-
Structure and symmetry inform gating principles of ionotropic glutamate receptors.Neuropharmacology. 2017 Jan;112(Pt A):11-15. doi: 10.1016/j.neuropharm.2016.08.034. Epub 2016 Sep 20. Neuropharmacology. 2017. PMID: 27663701 Free PMC article. Review.
-
Structural dynamics of an ionotropic glutamate receptor.Proteins. 2004 Aug 15;56(3):411-9. doi: 10.1002/prot.20154. Proteins. 2004. PMID: 15229875 Review.
-
Glycine agonism in ionotropic glutamate receptors.Neuropharmacology. 2021 Aug 1;193:108631. doi: 10.1016/j.neuropharm.2021.108631. Epub 2021 May 28. Neuropharmacology. 2021. PMID: 34058193 Review.
Cited by
-
On the molecular nature of large-pore channels.J Mol Biol. 2021 Aug 20;433(17):166994. doi: 10.1016/j.jmb.2021.166994. Epub 2021 Apr 16. J Mol Biol. 2021. PMID: 33865869 Free PMC article. Review.
-
A structurally derived model of subunit-dependent NMDA receptor function.J Physiol. 2018 Sep;596(17):4057-4089. doi: 10.1113/JP276093. Epub 2018 Aug 1. J Physiol. 2018. PMID: 29917241 Free PMC article.
-
Advancing NMDA Receptor Physiology by Integrating Multiple Approaches.Trends Neurosci. 2017 Mar;40(3):129-137. doi: 10.1016/j.tins.2017.01.001. Epub 2017 Feb 8. Trends Neurosci. 2017. PMID: 28187950 Free PMC article. Review.
-
Functional Evaluation of a De Novo GRIN2A Mutation Identified in a Patient with Profound Global Developmental Delay and Refractory Epilepsy.Mol Pharmacol. 2017 Apr;91(4):317-330. doi: 10.1124/mol.116.106781. Epub 2017 Jan 26. Mol Pharmacol. 2017. PMID: 28126851 Free PMC article.
-
Structural dynamics of GluK2 kainate receptors in apo and partial agonist bound states.Res Sq [Preprint]. 2023 Dec 2:rs.3.rs-3592604. doi: 10.21203/rs.3.rs-3592604/v1. Res Sq. 2023. PMID: 38076992 Free PMC article. Preprint.
References
-
- Hayashi T. Effects of sodium glutamate on the nervous system. Keio J. Med. 1954;3:192–193.
-
- Moriyoshi K, et al. Molecular cloning and characterization of the rat NMDA receptor. Nature. 1991;354:31–37. - PubMed
-
- Hollmann M, et al. Cloning by functional expression of a member of the glutamate receptor family. Nature. 1989;342:643–648. - PubMed
-
- Werner P, et al. Cloning of a putative high-affinity kainate receptor expressed predominantly in hippocampal CA3 cells. Nature. 1991;351:742–744. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

