Mutational Analysis of the Chlamydia muridarum Plasticity Zone
- PMID: 25939505
- PMCID: PMC4468545
- DOI: 10.1128/IAI.00106-15
Mutational Analysis of the Chlamydia muridarum Plasticity Zone
Abstract
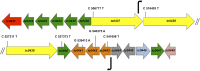
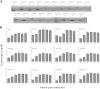
Pathogenically diverse Chlamydia spp. can have surprisingly similar genomes. Chlamydia trachomatis isolates that cause trachoma, sexually transmitted genital tract infections (chlamydia), and invasive lymphogranuloma venereum (LGV) and the murine strain Chlamydia muridarum share 99% of their gene content. A region of high genomic diversity between Chlamydia spp. termed the plasticity zone (PZ) may encode niche-specific virulence determinants that dictate pathogenic diversity. We hypothesized that PZ genes might mediate the greater virulence and gamma interferon (IFN-γ) resistance of C. muridarum compared to C. trachomatis in the murine genital tract. To test this hypothesis, we isolated and characterized a series of C. muridarum PZ nonsense mutants. Strains with nonsense mutations in chlamydial cytotoxins, guaBA-add, and a phospholipase D homolog developed normally in cell culture. Two of the cytotoxin mutants were less cytotoxic than the wild type, suggesting that the cytotoxins may be functional. However, none of the PZ nonsense mutants exhibited increased IFN-γ sensitivity in cell culture or were profoundly attenuated in a murine genital tract infection model. Our results suggest that C. muridarum PZ genes are transcribed--and some may produce functional proteins--but are dispensable for infection of the murine genital tract.
Copyright © 2015, American Society for Microbiology. All Rights Reserved.
Figures
Similar articles
-
Chlamydia muridarum Genital and Gastrointestinal Infection Tropism Is Mediated by Distinct Chromosomal Factors.Infect Immun. 2018 Jun 21;86(7):e00141-18. doi: 10.1128/IAI.00141-18. Print 2018 Jul. Infect Immun. 2018. PMID: 29661932 Free PMC article.
-
Inter-species lateral gene transfer focused on the Chlamydia plasticity zone identifies loci associated with immediate cytotoxicity and inclusion stability.Mol Microbiol. 2021 Dec;116(6):1433-1448. doi: 10.1111/mmi.14832. Epub 2021 Nov 16. Mol Microbiol. 2021. PMID: 34738268 Free PMC article.
-
A Genital Infection-Attenuated Chlamydia muridarum Mutant Infects the Gastrointestinal Tract and Protects against Genital Tract Challenge.mBio. 2020 Nov 3;11(6):e02770-20. doi: 10.1128/mBio.02770-20. mBio. 2020. PMID: 33144378 Free PMC article.
-
Chlamydia Spreading from the Genital Tract to the Gastrointestinal Tract - A Two-Hit Hypothesis.Trends Microbiol. 2018 Jul;26(7):611-623. doi: 10.1016/j.tim.2017.12.002. Epub 2017 Dec 27. Trends Microbiol. 2018. PMID: 29289422 Free PMC article. Review.
-
Chlamydial Plasmid-Dependent Pathogenicity.Trends Microbiol. 2017 Feb;25(2):141-152. doi: 10.1016/j.tim.2016.09.006. Epub 2016 Oct 3. Trends Microbiol. 2017. PMID: 27712952 Free PMC article. Review.
Cited by
-
Beyond Tryptophan Synthase: Identification of Genes That Contribute to Chlamydia trachomatis Survival during Gamma Interferon-Induced Persistence and Reactivation.Infect Immun. 2016 Sep 19;84(10):2791-801. doi: 10.1128/IAI.00356-16. Print 2016 Oct. Infect Immun. 2016. PMID: 27430273 Free PMC article.
-
The Impact of Lateral Gene Transfer in Chlamydia.Front Cell Infect Microbiol. 2022 Mar 7;12:861899. doi: 10.3389/fcimb.2022.861899. eCollection 2022. Front Cell Infect Microbiol. 2022. PMID: 35321311 Free PMC article. Review.
-
Phylogenetic analysis of human Chlamydia pneumoniae strains reveals a distinct Australian indigenous clade that predates European exploration of the continent.BMC Genomics. 2015 Dec 22;16:1094. doi: 10.1186/s12864-015-2281-y. BMC Genomics. 2015. PMID: 26694618 Free PMC article.
-
Bringing genetics to heretofore intractable obligate intracellular bacterial pathogens: Chlamydia and beyond.PLoS Pathog. 2022 Jul 28;18(7):e1010669. doi: 10.1371/journal.ppat.1010669. eCollection 2022 Jul. PLoS Pathog. 2022. PMID: 35901011 Free PMC article. No abstract available.
-
From genomes to genotypes: molecular epidemiological analysis of Chlamydia gallinacea reveals a high level of genetic diversity for this newly emerging chlamydial pathogen.BMC Genomics. 2017 Dec 6;18(1):949. doi: 10.1186/s12864-017-4343-9. BMC Genomics. 2017. PMID: 29212448 Free PMC article.
References
-
- Read TD, Brunham RC, Shen C, Gill SR, Heidelberg JF, White O, Hickey EK, Peterson J, Utterback T, Berry K, Bass S, Linher K, Weidman J, Khouri H, Craven B, Bowman C, Dodson R, Gwinn M, Nelson W, DeBoy R, Kolonay J, McClarty G, Salzberg SL, Eisen J, Fraser CM. 2000. Genome sequences of Chlamydia trachomatis MoPn and Chlamydia pneumoniae AR39. Nucleic Acids Res 28:1397–1406. doi:10.1093/nar/28.6.1397. - DOI - PMC - PubMed
-
- Read TD, Myers GSA, Brunham RC, Nelson WC, Paulsen IT, Heidelberg J, Holtzapple E, Khouri H, Federova NB, Carty HA, Umayam LA, Haft DH, Peterson J, Beanan MJ, White O, Salzberg SL, Hsia RC, McClarty G, Rank RG, Bavoil PM, Fraser CM. 2003. Genome sequence of Chlamydophila caviae (Chlamydia psittaci GPIC): examining the role of niche-specific genes in the evolution of the Chlamydiaceae. Nucleic Acids Res 31:2134–2147. doi:10.1093/nar/gkg321. - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Miscellaneous

