Whole Genomic Analysis of Human G12P[6] and G12P[8] Rotavirus Strains that Have Emerged in Myanmar
- PMID: 25938434
- PMCID: PMC4418666
- DOI: 10.1371/journal.pone.0124965
Whole Genomic Analysis of Human G12P[6] and G12P[8] Rotavirus Strains that Have Emerged in Myanmar
Abstract
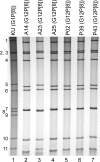
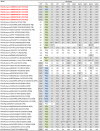
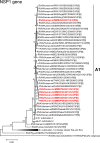
G12 rotaviruses are emerging rotavirus strains causing severe diarrhea in infants and young children worldwide. However, the whole genomes of only a few G12 strains have been fully sequenced and analyzed. In this study, we sequenced and characterized the complete genomes of six G12 strains (RVA/Human-tc/MMR/A14/2011/G12P[8], RVA/Human-tc/MMR/A23/2011/G12P[6], RVA/Human-tc/MMR/A25/2011/G12P[8], RVA/Human-tc/MMR/P02/2011/G12P[8], RVA/Human-tc/MMR/P39/2011/G12P[8], and RVA/Human-tc/MMR/P43/2011/G12P[8]) detected in six stool samples from children with acute gastroenteritis in Myanmar. On whole genomic analysis, all six Myanmarese G12 strains were found to have a Wa-like genetic backbone: G12-P[8]-I1-R1-C1-M1-A1-N1-T1-E1-H1 for strains A14, A25, P02, P39, and P43, and G12-P[6]-I1-R1-C1-M1-A1-N1-T1-E1-H1 for strain A23. Phylogenetic analysis showed that most genes of the six strains examined in this study were genetically related to globally circulating human G1, G3, G9, and G12 strains. Of note is that the NSP4 gene of strain A23 exhibited the closest relationship with the cognate genes of human-like bovine strains as well as human strains, suggesting the occurrence of reassortment between human and bovine strains. Furthermore, strains A14, A25, P02, P39, and P43 were very closely related to one another in all the 11 gene segments, indicating derivation of the five strains from a common origin. On the other hand, strain A23 consistently formed distinct clusters as to all the 11 gene segments, indicating a distinct origin of strain A23 from that of strains A14, A25, P02, P39, and P43. To our knowledge, this is the first report on whole genome-based characterization of G12 strains that have emerged in Myanmar. Our observations will provide important insights into the evolutionary dynamics of spreading G12 rotaviruses in Asia.
Conflict of interest statement
Figures










Similar articles
-
Whole Genome Analysis of African G12P[6] and G12P[8] Rotaviruses Provides Evidence of Porcine-Human Reassortment at NSP2, NSP3, and NSP4.Front Microbiol. 2021 Jan 12;11:604444. doi: 10.3389/fmicb.2020.604444. eCollection 2020. Front Microbiol. 2021. PMID: 33510725 Free PMC article.
-
Whole genomic analysis of human G12P[6] and G12P[8] rotavirus strains that have emerged in Kenya: identification of porcine-like NSP4 genes.Infect Genet Evol. 2014 Oct;27:277-93. doi: 10.1016/j.meegid.2014.08.002. Epub 2014 Aug 8. Infect Genet Evol. 2014. PMID: 25111611
-
Whole genome analyses of African G2, G8, G9, and G12 rotavirus strains using sequence-independent amplification and 454® pyrosequencing.J Med Virol. 2011 Nov;83(11):2018-42. doi: 10.1002/jmv.22207. J Med Virol. 2011. PMID: 21915879
-
Whole-genome characterisation of G12P[6] rotavirus strains possessing two distinct genotype constellations co-circulating in Blantyre, Malawi, 2008.Arch Virol. 2017 Jan;162(1):213-226. doi: 10.1007/s00705-016-3103-5. Epub 2016 Oct 7. Arch Virol. 2017. PMID: 27718073 Clinical Trial.
-
Genotype constellation and evolution of group A rotaviruses infecting humans.Curr Opin Virol. 2012 Aug;2(4):426-33. doi: 10.1016/j.coviro.2012.04.007. Epub 2012 Jun 9. Curr Opin Virol. 2012. PMID: 22683209 Review.
Cited by
-
Molecular characterization of human group A rotavirus genotypes circulating in Rawalpindi, Islamabad, Pakistan during 2015-2016.PLoS One. 2019 Jul 30;14(7):e0220387. doi: 10.1371/journal.pone.0220387. eCollection 2019. PLoS One. 2019. PMID: 31361761 Free PMC article.
-
Porcine Rotaviruses: Epidemiology, Immune Responses and Control Strategies.Viruses. 2017 Mar 18;9(3):48. doi: 10.3390/v9030048. Viruses. 2017. PMID: 28335454 Free PMC article. Review.
-
Whole-Genome Analyses Identifies Multiple Reassortant Rotavirus Strains in Rwanda Post-Vaccine Introduction.Viruses. 2021 Jan 12;13(1):95. doi: 10.3390/v13010095. Viruses. 2021. PMID: 33445703 Free PMC article.
-
Reassortment of Human and Animal Rotavirus Gene Segments in Emerging DS-1-Like G1P[8] Rotavirus Strains.PLoS One. 2016 Feb 4;11(2):e0148416. doi: 10.1371/journal.pone.0148416. eCollection 2016. PLoS One. 2016. PMID: 26845439 Free PMC article.
-
Whole Genome Analysis of African G12P[6] and G12P[8] Rotaviruses Provides Evidence of Porcine-Human Reassortment at NSP2, NSP3, and NSP4.Front Microbiol. 2021 Jan 12;11:604444. doi: 10.3389/fmicb.2020.604444. eCollection 2020. Front Microbiol. 2021. PMID: 33510725 Free PMC article.
References
-
- Tate JE, Burton AH, Boschi-Pinto C, Steele AD, Dugue J, Parashar UD, et al. 2008 estimate of worldwide rotavirus-associated mortality in children younger than 5 years before the introduction of universal rotavirus vaccination programmes: a systemic review and meta-analysis. Lancet Infect Dis. 2012;12: 136–141. 10.1016/S1473-3099(11)70253-5 - DOI - PubMed
-
- Kahn G, Fitzwater S, Tate J, Kang G, Ganguly N, Nair G, et al. Epidemiology and prospects for prevention of rotavirus disease in India. Indian Pediatr. 2012;49: 467–474. - PubMed
-
- Moe K, Hummelman EG, Oo WM, Lwin T, Htwe TT. Hospital-based surveillance for rotavirus diarrhea in children in Yangon, Myanmar. J Infect Dis. 2005;192 Suppl 1: S111–S113. - PubMed
-
- Estes MK, Greenberg HB. Rotaviruses In: Knipe DM, Howley PM, editors. Fields Virology. 6th ed. Philadelphia: Lippincott Williams & Wilkins; 2013. pp. 1347–1401.
Publication types
MeSH terms
Substances
Associated data
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

