A unique chromatin complex occupies young α-satellite arrays of human centromeres
- PMID: 25927077
- PMCID: PMC4410388
- DOI: 10.1126/sciadv.1400234
A unique chromatin complex occupies young α-satellite arrays of human centromeres
Abstract
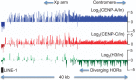
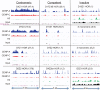
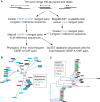
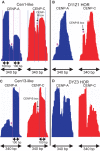
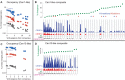
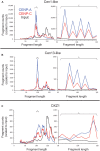
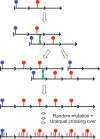
The intractability of homogeneous α-satellite arrays has impeded understanding of human centromeres. Artificial centromeres are produced from higher-order repeats (HORs) present at centromere edges, although the exact sequences and chromatin conformations of centromere cores remain unknown. We use high-resolution chromatin immunoprecipitation (ChIP) of centromere components followed by clustering of sequence data as an unbiased approach to identify functional centromere sequences. We find that specific dimeric α-satellite units shared by multiple individuals dominate functional human centromeres. We identify two recently homogenized α-satellite dimers that are occupied by precisely positioned CENP-A (cenH3) nucleosomes with two ~100-base pair (bp) DNA wraps in tandem separated by a CENP-B/CENP-C-containing linker, whereas pericentromeric HORs show diffuse positioning. Precise positioning is largely maintained, whereas abundance decreases exponentially with divergence, which suggests that young α-satellite dimers with paired ~100-bp particles mediate evolution of functional human centromeres. Our unbiased strategy for identifying functional centromeric sequences should be generally applicable to tandem repeat arrays that dominate the centromeres of most eukaryotes.
Figures


Similar articles
-
DNA satellite and chromatin organization at mouse centromeres and pericentromeres.Genome Biol. 2024 Feb 20;25(1):52. doi: 10.1186/s13059-024-03184-z. Genome Biol. 2024. PMID: 38378611 Free PMC article.
-
Remarkable Evolutionary Plasticity of Centromeric Chromatin.Cold Spring Harb Symp Quant Biol. 2017;82:71-82. doi: 10.1101/sqb.2017.82.033605. Epub 2017 Dec 1. Cold Spring Harb Symp Quant Biol. 2017. PMID: 29196559
-
Repeatless and repeat-based centromeres in potato: implications for centromere evolution.Plant Cell. 2012 Sep;24(9):3559-74. doi: 10.1105/tpc.112.100511. Epub 2012 Sep 11. Plant Cell. 2012. PMID: 22968715 Free PMC article.
-
Structural and functional dynamics of human centromeric chromatin.Annu Rev Genomics Hum Genet. 2006;7:301-13. doi: 10.1146/annurev.genom.7.080505.115613. Annu Rev Genomics Hum Genet. 2006. PMID: 16756479 Review.
-
What makes a centromere?Exp Cell Res. 2020 Apr 15;389(2):111895. doi: 10.1016/j.yexcr.2020.111895. Epub 2020 Feb 6. Exp Cell Res. 2020. PMID: 32035948 Review.
Cited by
-
The genetics and epigenetics of satellite centromeres.Genome Res. 2022 Apr;32(4):608-615. doi: 10.1101/gr.275351.121. Epub 2022 Mar 31. Genome Res. 2022. PMID: 35361623 Free PMC article.
-
The Dynamic Structure and Rapid Evolution of Human Centromeric Satellite DNA.Genes (Basel). 2022 Dec 28;14(1):92. doi: 10.3390/genes14010092. Genes (Basel). 2022. PMID: 36672831 Free PMC article. Review.
-
The cellular mechanisms and consequences of centromere drive.Curr Opin Cell Biol. 2018 Jun;52:58-65. doi: 10.1016/j.ceb.2018.01.011. Epub 2018 Feb 16. Curr Opin Cell Biol. 2018. PMID: 29454259 Free PMC article. Review.
-
Molecular Complexes at Euchromatin, Heterochromatin and Centromeric Chromatin.Int J Mol Sci. 2021 Jun 28;22(13):6922. doi: 10.3390/ijms22136922. Int J Mol Sci. 2021. PMID: 34203193 Free PMC article. Review.
-
Stable centromere positioning in diverse sequence contexts of complex and satellite centromeres of maize and wild relatives.Genome Biol. 2017 Jun 21;18(1):121. doi: 10.1186/s13059-017-1249-4. Genome Biol. 2017. PMID: 28637491 Free PMC article.
References
-
- Alexandrov I., Kazakov A., Tumeneva I, Shepelev V., Yurov Y., Alpha-satellite DNA of primates: Old and new families. Chromosoma 110, 253–266 (2001). - PubMed
-
- Schueler M. G., Higgins A. W., Rudd M. K., Gustashaw K., Willard H. F., Genomic and genetic definition of a functional human centromere. Science 294, 109–115 (2001). - PubMed
-
- Melters D. P., Bradnam K. R., Young H. A., Telis N., May M. R., Ruby J. G., Sebra R., Peluso P., Eid J., Rank D., Garcia J. F., DeRisi J. L., Smith T., Tobias C., Ross-Ibarra J., Korf I., Chan S. W. L., Comparative analysis of tandem repeats from hundreds of species reveals unique insights into centromere evolution. Genome Biol. 14, R10 (2013). - PMC - PubMed
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

