Neddylation promotes ubiquitylation and release of Ku from DNA-damage sites
- PMID: 25921528
- PMCID: PMC4431666
- DOI: 10.1016/j.celrep.2015.03.058
Neddylation promotes ubiquitylation and release of Ku from DNA-damage sites
Abstract
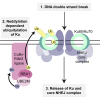
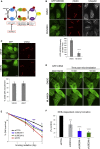
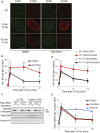
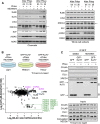
The activities of many DNA-repair proteins are controlled through reversible covalent modification by ubiquitin and ubiquitin-like molecules. Nonhomologous end-joining (NHEJ) is the predominant DNA double-strand break (DSB) repair pathway in mammalian cells and is initiated by DSB ends being recognized by the Ku70/Ku80 (Ku) heterodimer. By using MLN4924, an anti-cancer drug in clinical trials that specifically inhibits conjugation of the ubiquitin-like protein, NEDD8, to target proteins, we demonstrate that NEDD8 accumulation at DNA-damage sites is a highly dynamic process. In addition, we show that depleting cells of the NEDD8 E2-conjugating enzyme, UBE2M, yields ionizing radiation hypersensitivity and reduced cell survival following NHEJ. Finally, we demonstrate that neddylation promotes Ku ubiquitylation after DNA damage and release of Ku and Ku-associated proteins from damage sites following repair. These studies provide insights into how the NHEJ core complex dissociates from repair sites and highlight its importance for cell survival following DSB induction.
Copyright © 2015 The Authors. Published by Elsevier Inc. All rights reserved.
Figures

Similar articles
-
The deubiquitylating enzyme UCHL3 regulates Ku80 retention at sites of DNA damage.Sci Rep. 2018 Dec 17;8(1):17891. doi: 10.1038/s41598-018-36235-0. Sci Rep. 2018. PMID: 30559450 Free PMC article.
-
Mycobacterium tuberculosis Ku can bind to nuclear DNA damage and sensitize mammalian cells to bleomycin sulfate.Mutagenesis. 2011 Nov;26(6):795-803. doi: 10.1093/mutage/ger049. Epub 2011 Aug 2. Mutagenesis. 2011. PMID: 21811007 Free PMC article.
-
An SCF complex containing Fbxl12 mediates DNA damage-induced Ku80 ubiquitylation.Cell Cycle. 2013 Feb 15;12(4):587-95. doi: 10.4161/cc.23408. Epub 2013 Jan 16. Cell Cycle. 2013. PMID: 23324393 Free PMC article.
-
The Ku heterodimer: function in DNA repair and beyond.Mutat Res Rev Mutat Res. 2015 Jan-Mar;763:15-29. doi: 10.1016/j.mrrev.2014.06.002. Epub 2014 Jul 4. Mutat Res Rev Mutat Res. 2015. PMID: 25795113 Review.
-
Destroying the ring: Freeing DNA from Ku with ubiquitin.FEBS Lett. 2011 Sep 16;585(18):2876-82. doi: 10.1016/j.febslet.2011.05.046. Epub 2011 Jun 1. FEBS Lett. 2011. PMID: 21640108 Free PMC article. Review.
Cited by
-
Direct Conjugation of NEDD8 to the N-Terminus of a Model Protein Can Induce Degradation.Cells. 2021 Apr 9;10(4):854. doi: 10.3390/cells10040854. Cells. 2021. PMID: 33918652 Free PMC article.
-
UBE2M promotes cell proliferation via the β-catenin/cyclin D1 signaling in hepatocellular carcinoma.Aging (Albany NY). 2020 Feb 3;12(3):2373-2392. doi: 10.18632/aging.102749. Epub 2020 Feb 3. Aging (Albany NY). 2020. PMID: 32012120 Free PMC article.
-
Structure and mechanism in non-homologous end joining.DNA Repair (Amst). 2023 Oct;130:103547. doi: 10.1016/j.dnarep.2023.103547. Epub 2023 Jul 29. DNA Repair (Amst). 2023. PMID: 37556875 Free PMC article.
-
Mutational phospho-mimicry reveals a regulatory role for the XRCC4 and XLF C-terminal tails in modulating DNA bridging during classical non-homologous end joining.Elife. 2017 May 13;6:e22900. doi: 10.7554/eLife.22900. Elife. 2017. PMID: 28500754 Free PMC article.
-
How cells ensure correct repair of DNA double-strand breaks.J Biol Chem. 2018 Jul 6;293(27):10502-10511. doi: 10.1074/jbc.TM118.000371. Epub 2018 Feb 5. J Biol Chem. 2018. PMID: 29414795 Free PMC article. Review.
References
-
- Brownell J.E., Sintchak M.D., Gavin J.M., Liao H., Bruzzese F.J., Bump N.J., Soucy T.A., Milhollen M.A., Yang X., Burkhardt A.L. Substrate-assisted inhibition of ubiquitin-like protein-activating enzymes: the NEDD8 E1 inhibitor MLN4924 forms a NEDD8-AMP mimetic in situ. Mol. Cell. 2010;37:102–111. - PubMed
-
- Cope G.A., Suh G.S., Aravind L., Schwarz S.E., Zipursky S.L., Koonin E.V., Deshaies R.J. Role of predicted metalloprotease motif of Jab1/Csn5 in cleavage of Nedd8 from Cul1. Science. 2002;298:608–611. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials
Miscellaneous

