A Cdh1-APC/FMRP Ubiquitin Signaling Link Drives mGluR-Dependent Synaptic Plasticity in the Mammalian Brain
- PMID: 25913861
- PMCID: PMC4492466
- DOI: 10.1016/j.neuron.2015.03.049
A Cdh1-APC/FMRP Ubiquitin Signaling Link Drives mGluR-Dependent Synaptic Plasticity in the Mammalian Brain
Abstract
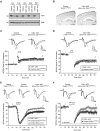
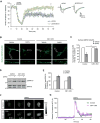
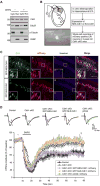
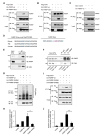
Deregulation of synaptic plasticity may contribute to the pathogenesis of developmental cognitive disorders. In particular, exaggerated mGluR-dependent LTD is featured in fragile X syndrome, but the mechanisms that regulate mGluR-LTD remain incompletely understood. We report that conditional knockout of Cdh1, the key regulatory subunit of the ubiquitin ligase Cdh1-anaphase-promoting complex (Cdh1-APC), profoundly impairs mGluR-LTD in the hippocampus. Mechanistically, we find that Cdh1-APC operates in the cytoplasm to drive mGluR-LTD. We also identify the fragile X syndrome protein FMRP as a substrate of Cdh1-APC. Endogenous Cdh1-APC forms a complex with endogenous FMRP, and knockout of Cdh1 impairs mGluR-induced ubiquitination and degradation of FMRP in the hippocampus. Knockout of FMRP suppresses, and expression of an FMRP mutant protein that fails to interact with Cdh1 phenocopies, the Cdh1 knockout phenotype of impaired mGluR-LTD. These findings define Cdh1-APC and FMRP as components of a novel ubiquitin signaling pathway that regulates mGluR-LTD in the brain.
Copyright © 2015 Elsevier Inc. All rights reserved.
Figures
Similar articles
-
Dynamic translational and proteasomal regulation of fragile X mental retardation protein controls mGluR-dependent long-term depression.Neuron. 2006 Aug 17;51(4):441-54. doi: 10.1016/j.neuron.2006.07.005. Neuron. 2006. PMID: 16908410
-
Altered hippocampal synaptic plasticity in the FMR1 gene family knockout mouse models.J Neurophysiol. 2009 May;101(5):2572-80. doi: 10.1152/jn.90558.2008. Epub 2009 Feb 25. J Neurophysiol. 2009. PMID: 19244359 Free PMC article.
-
Loss of the fragile X mental retardation protein decouples metabotropic glutamate receptor dependent priming of long-term potentiation from protein synthesis.J Neurophysiol. 2010 Aug;104(2):1047-51. doi: 10.1152/jn.00449.2010. Epub 2010 Jun 16. J Neurophysiol. 2010. PMID: 20554840 Free PMC article.
-
Metabotropic glutamate receptors and fragile x mental retardation protein: partners in translational regulation at the synapse.Sci Signal. 2008 Feb 5;1(5):pe6. doi: 10.1126/stke.15pe6. Sci Signal. 2008. PMID: 18272470 Review.
-
The mGluR theory of fragile X mental retardation.Trends Neurosci. 2004 Jul;27(7):370-7. doi: 10.1016/j.tins.2004.04.009. Trends Neurosci. 2004. PMID: 15219735 Review.
Cited by
-
Ubiquitin and Ubiquitin-Like Proteins in the Critical Equilibrium between Synapse Physiology and Intellectual Disability.eNeuro. 2020 Aug 26;7(4):ENEURO.0137-20.2020. doi: 10.1523/ENEURO.0137-20.2020. Print 2020 Jul/Aug. eNeuro. 2020. PMID: 32719102 Free PMC article. Review.
-
Oppositional poly(A) tail length regulation by FMRP and CPEB1.RNA. 2022 May;28(5):756-765. doi: 10.1261/rna.079050.121. Epub 2022 Feb 25. RNA. 2022. PMID: 35217597 Free PMC article.
-
Chronic Activation of Gp1 mGluRs Leads to Distinct Refinement of Neural Network Activity through Non-Canonical p53 and Akt Signaling.eNeuro. 2020 Mar 27;7(2):ENEURO.0438-19.2020. doi: 10.1523/ENEURO.0438-19.2020. Print 2020 Mar/Apr. eNeuro. 2020. PMID: 32161037 Free PMC article.
-
The ubiquitin-proteasome system functionally links neuronal Tomosyn-1 to dendritic morphology.J Biol Chem. 2018 Feb 16;293(7):2232-2246. doi: 10.1074/jbc.M117.815514. Epub 2017 Dec 21. J Biol Chem. 2018. PMID: 29269412 Free PMC article.
-
Synaptic Plasticity, a Prominent Contributor to the Anxiety in Fragile X Syndrome.Neural Plast. 2016;2016:9353929. doi: 10.1155/2016/9353929. Epub 2016 Apr 28. Neural Plast. 2016. PMID: 27239350 Free PMC article. Review.
References
-
- Bear MF, Huber KM, Warren ST. The mGluR theory of fragile X mental retardation. Trends in neurosciences. 2004;27:370–377. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Miscellaneous

