The genomes of two key bumblebee species with primitive eusocial organization
- PMID: 25908251
- PMCID: PMC4414376
- DOI: 10.1186/s13059-015-0623-3
The genomes of two key bumblebee species with primitive eusocial organization
Abstract
Background: The shift from solitary to social behavior is one of the major evolutionary transitions. Primitively eusocial bumblebees are uniquely placed to illuminate the evolution of highly eusocial insect societies. Bumblebees are also invaluable natural and agricultural pollinators, and there is widespread concern over recent population declines in some species. High-quality genomic data will inform key aspects of bumblebee biology, including susceptibility to implicated population viability threats.
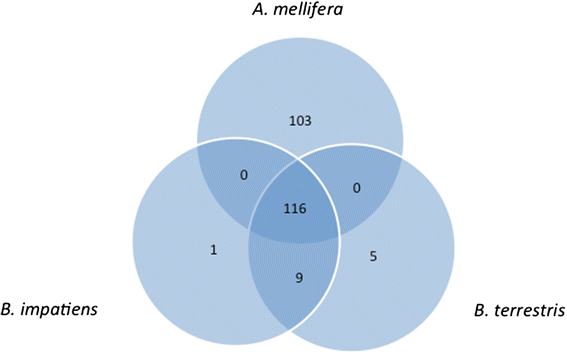
Results: We report the high quality draft genome sequences of Bombus terrestris and Bombus impatiens, two ecologically dominant bumblebees and widely utilized study species. Comparing these new genomes to those of the highly eusocial honeybee Apis mellifera and other Hymenoptera, we identify deeply conserved similarities, as well as novelties key to the biology of these organisms. Some honeybee genome features thought to underpin advanced eusociality are also present in bumblebees, indicating an earlier evolution in the bee lineage. Xenobiotic detoxification and immune genes are similarly depauperate in bumblebees and honeybees, and multiple categories of genes linked to social organization, including development and behavior, show high conservation. Key differences identified include a bias in bumblebee chemoreception towards gustation from olfaction, and striking differences in microRNAs, potentially responsible for gene regulation underlying social and other traits.
Conclusions: These two bumblebee genomes provide a foundation for post-genomic research on these key pollinators and insect societies. Overall, gene repertoires suggest that the route to advanced eusociality in bees was mediated by many small changes in many genes and processes, and not by notable expansion or depauperation.
Figures
Comment in
-
The making of eusociality: insights from two bumblebee genomes.Genome Biol. 2015 Apr 24;16(1):75. doi: 10.1186/s13059-015-0635-z. Genome Biol. 2015. PMID: 25908513 Free PMC article.
Similar articles
-
The nuclear and mitochondrial genomes of Frieseomelitta varia - a highly eusocial stingless bee (Meliponini) with a permanently sterile worker caste.BMC Genomics. 2020 Jun 3;21(1):386. doi: 10.1186/s12864-020-06784-8. BMC Genomics. 2020. PMID: 32493270 Free PMC article.
-
A depauperate immune repertoire precedes evolution of sociality in bees.Genome Biol. 2015 Apr 24;16(1):83. doi: 10.1186/s13059-015-0628-y. Genome Biol. 2015. PMID: 25908406 Free PMC article.
-
Molecular heterochrony and the evolution of sociality in bumblebees (Bombus terrestris).Proc Biol Sci. 2014 Feb 19;281(1780):20132419. doi: 10.1098/rspb.2013.2419. Print 2014 Apr 7. Proc Biol Sci. 2014. PMID: 24552837 Free PMC article.
-
Insights into the molecular basis of social behaviour from studies on the honeybee, Apis mellifera.Invert Neurosci. 2008 Mar;8(1):1-9. doi: 10.1007/s10158-008-0066-6. Epub 2008 Feb 15. Invert Neurosci. 2008. PMID: 18274798 Review.
-
Bumblebees with the socially transmitted microbiome: A novel model organism for gut microbiota research.Insect Sci. 2022 Aug;29(4):958-976. doi: 10.1111/1744-7917.13040. Epub 2022 May 14. Insect Sci. 2022. PMID: 35567381 Review.
Cited by
-
Signals of adaptation to agricultural stress in the genomes of two European bumblebees.Front Genet. 2022 Oct 5;13:993416. doi: 10.3389/fgene.2022.993416. eCollection 2022. Front Genet. 2022. PMID: 36276969 Free PMC article.
-
Evolution of odorant receptor repertoires across Hymenoptera is not linked to the evolution of eusociality.Proc Biol Sci. 2024 Sep;291(2031):20241280. doi: 10.1098/rspb.2024.1280. Epub 2024 Sep 25. Proc Biol Sci. 2024. PMID: 39317325 Free PMC article.
-
Substances in the mandibular glands mediate queen effects on larval development and colony organization in an annual bumble bee.Proc Natl Acad Sci U S A. 2023 Nov 7;120(45):e2302071120. doi: 10.1073/pnas.2302071120. Epub 2023 Oct 30. Proc Natl Acad Sci U S A. 2023. PMID: 37903277 Free PMC article.
-
Identification and functional characterisation of a novel N-cyanoamidine neonicotinoid metabolising cytochrome P450, CYP9Q6, from the buff-tailed bumblebee Bombus terrestris.Insect Biochem Mol Biol. 2019 Aug;111:103171. doi: 10.1016/j.ibmb.2019.05.006. Epub 2019 May 25. Insect Biochem Mol Biol. 2019. PMID: 31136794 Free PMC article.
-
Pollinator parasites and the evolution of floral traits.Ecol Evol. 2019 May 15;9(11):6722-6737. doi: 10.1002/ece3.4989. eCollection 2019 Jun. Ecol Evol. 2019. PMID: 31236255 Free PMC article.
References
-
- Maynard Smith J, Szathmary E. The major evolutionary transitions. Nature. 1995;374:227–232. - PubMed
-
- Crozier R, Pamilo P. Evolution of social insect colonies: Sex allocation and kin selection. Oxford: Oxford University Press; 1996.
-
- Hughes WO, Oldroyd BP, Beekman M, Ratnieks FL. Ancestral monogamy shows kin selection is key to the evolution of eusociality. Science. 2008;320:1213–1216. - PubMed
-
- Hamilton WD. The genetical evolution of social behaviour I. J Theor Biol. 1964;7:1–16. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

