Antibodies to the core proteins of Nairobi sheep disease virus/Ganjam virus reveal details of the distribution of the proteins in infected cells and tissues
- PMID: 25905707
- PMCID: PMC4407892
- DOI: 10.1371/journal.pone.0124966
Antibodies to the core proteins of Nairobi sheep disease virus/Ganjam virus reveal details of the distribution of the proteins in infected cells and tissues
Abstract
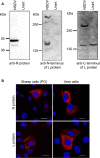
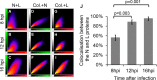
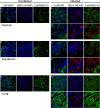
Nairobi sheep disease virus (NSDV; also called Ganjam virus in India) is a bunyavirus of the genus Nairovirus. It causes a haemorrhagic gastroenteritis in sheep and goats with mortality up to 90%. The virus is closely related to the human pathogen Crimean-Congo haemorrhagic fever virus (CCHFV). Little is currently known about the biology of NSDV. We have generated specific antibodies against the virus nucleocapsid protein (N) and polymerase (L) and used these to characterise NSDV in infected cells and to study its distribution during infection in a natural host. Due to its large size and the presence of a papain-like protease (the OTU-like domain) it has been suggested that the L protein of nairoviruses undergoes an autoproteolytic cleavage into polymerase and one or more accessory proteins. Specific antibodies which recognise either the N-terminus or the C-terminus of the NSDV L protein showed no evidence of L protein cleavage in NSDV-infected cells. Using the specific anti-N and anti-L antibodies, it was found that these viral proteins do not fully colocalise in infected cells; the N protein accumulated near the Golgi at early stages of infection while the L protein was distributed throughout the cytoplasm, further supporting the multifunctional nature of the L protein. These antibodies also allowed us to gain information about the organs and cell types targeted by the virus in vivo. We could detect NSDV in cryosections prepared from various tissues collected post-mortem from experimentally inoculated animals; the virus was found in the mucosal lining of the small and large intestine, in the lungs, and in mesenteric lymph nodes (MLN), where NSDV appeared to target monocytes and/or macrophages.
Conflict of interest statement
Figures

Similar articles
-
Generation and Characterisation of Monoclonal Antibodies against Nairobi Sheep Disease Virus Nucleoprotein.Viruses. 2023 Sep 5;15(9):1876. doi: 10.3390/v15091876. Viruses. 2023. PMID: 37766282 Free PMC article.
-
The nairovirus nairobi sheep disease virus/ganjam virus induces the translocation of protein disulphide isomerase-like oxidoreductases from the endoplasmic reticulum to the cell surface and the extracellular space.PLoS One. 2014 Apr 8;9(4):e94656. doi: 10.1371/journal.pone.0094656. eCollection 2014. PLoS One. 2014. PMID: 24714576 Free PMC article.
-
Nairobi sheep disease virus/Ganjam virus.Rev Sci Tech. 2015 Aug;34(2):411-7. doi: 10.20506/rst.34.2.2367. Rev Sci Tech. 2015. PMID: 26647464
-
Ganjam virus.Indian J Med Res. 2009 Nov;130(5):514-9. Indian J Med Res. 2009. PMID: 20090098 Review.
-
The molecular biology of nairoviruses, an emerging group of tick-borne arboviruses.Arch Virol. 2014 Jun;159(6):1249-65. doi: 10.1007/s00705-013-1940-z. Epub 2013 Dec 11. Arch Virol. 2014. PMID: 24327094 Free PMC article. Review.
Cited by
-
Generation and Characterisation of Monoclonal Antibodies against Nairobi Sheep Disease Virus Nucleoprotein.Viruses. 2023 Sep 5;15(9):1876. doi: 10.3390/v15091876. Viruses. 2023. PMID: 37766282 Free PMC article.
-
ISG15 overexpression compensates the defect of Crimean-Congo hemorrhagic fever virus polymerase bearing a protease-inactive ovarian tumor domain.PLoS Negl Trop Dis. 2020 Sep 15;14(9):e0008610. doi: 10.1371/journal.pntd.0008610. eCollection 2020 Sep. PLoS Negl Trop Dis. 2020. PMID: 32931521 Free PMC article.
-
The Integration of Human and Veterinary Studies for Better Understanding and Management of Crimean-Congo Haemorrhagic Fever.Front Immunol. 2021 Mar 18;12:629636. doi: 10.3389/fimmu.2021.629636. eCollection 2021. Front Immunol. 2021. PMID: 33815379 Free PMC article. Review.
-
The Adaptive Immune Response against Bunyavirales.Viruses. 2024 Mar 21;16(3):483. doi: 10.3390/v16030483. Viruses. 2024. PMID: 38543848 Free PMC article. Review.
References
-
- Casals J, Tignor GH. The Nairovirus genus: serological relationships. Intervirology. 1980;14(3–4):144–147. Epub 1980/01/01. PubMed . - PubMed
-
- Clerx JP, Casals J, Bishop DH. Structural characteristics of nairoviruses (genus Nairovirus, Bunyaviridae). J Gen Virol. 1981;55(Pt 1):165–178. Epub 1981/07/01. PubMed . - PubMed
-
- Zeller HG, Karabatsos N, Calisher CH, Digoutte JP, Cropp CB, Murphy FA, et al. Electron microscopic and antigenic studies of uncharacterized viruses. II. Evidence suggesting the placement of viruses in the family Bunyaviridae. Arch Virol. 1989;108(3–4):211–227. PubMed . - PubMed
-
- White WR. Nairobi Sheep Disease In: 7th, editor. Foreign Animal Disease: Boca Publications Group; 2008. p. 335–342.
-
- Uilenberg G. General review of tick-borne diseases of sheep and goats world-wide. Parassitologia. 1997;39(2):161–165. Epub 1997/06/01. PubMed . - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

