DNA damage response and spindle assembly checkpoint function throughout the cell cycle to ensure genomic integrity
- PMID: 25898113
- PMCID: PMC4405263
- DOI: 10.1371/journal.pgen.1005150
DNA damage response and spindle assembly checkpoint function throughout the cell cycle to ensure genomic integrity
Abstract
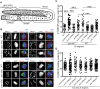
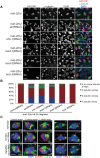
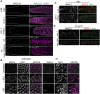
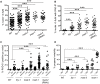
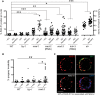
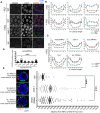
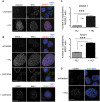
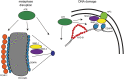
Errors in replication or segregation lead to DNA damage, mutations, and aneuploidies. Consequently, cells monitor these events and delay progression through the cell cycle so repair precedes division. The DNA damage response (DDR), which monitors DNA integrity, and the spindle assembly checkpoint (SAC), which responds to defects in spindle attachment/tension during metaphase of mitosis and meiosis, are critical for preventing genome instability. Here we show that the DDR and SAC function together throughout the cell cycle to ensure genome integrity in C. elegans germ cells. Metaphase defects result in enrichment of SAC and DDR components to chromatin, and both SAC and DDR are required for metaphase delays. During persistent metaphase arrest following establishment of bi-oriented chromosomes, stability of the metaphase plate is compromised in the absence of DDR kinases ATR or CHK1 or SAC components, MAD1/MAD2, suggesting SAC functions in metaphase beyond its interactions with APC activator CDC20. In response to DNA damage, MAD2 and the histone variant CENPA become enriched at the nuclear periphery in a DDR-dependent manner. Further, depletion of either MAD1 or CENPA results in loss of peripherally associated damaged DNA. In contrast to a SAC-insensitive CDC20 mutant, germ cells deficient for SAC or CENPA cannot efficiently repair DNA damage, suggesting that SAC mediates DNA repair through CENPA interactions with the nuclear periphery. We also show that replication perturbations result in relocalization of MAD1/MAD2 in human cells, suggesting that the role of SAC in DNA repair is conserved.
Conflict of interest statement
The authors have declared that no competing interests exist.
Figures
Similar articles
-
Working on Genomic Stability: From the S-Phase to Mitosis.Genes (Basel). 2020 Feb 20;11(2):225. doi: 10.3390/genes11020225. Genes (Basel). 2020. PMID: 32093406 Free PMC article. Review.
-
Caenorhabditis elegans BUB-3 and SAN-1/MAD3 Spindle Assembly Checkpoint Components Are Required for Genome Stability in Response to Treatment with Ionizing Radiation.G3 (Bethesda). 2017 Dec 4;7(12):3875-3885. doi: 10.1534/g3.117.1122. G3 (Bethesda). 2017. PMID: 29046436 Free PMC article.
-
CHK1-CENP B/MAD2 is associated with mild oxidative damage-induced sex chromosome aneuploidy of male mouse embryos during in vitro fertilization.Free Radic Biol Med. 2019 Jun;137:181-193. doi: 10.1016/j.freeradbiomed.2019.04.037. Epub 2019 Apr 28. Free Radic Biol Med. 2019. PMID: 31042615
-
Inhibition of CDK7 bypasses spindle assembly checkpoint via premature cyclin B degradation during oocyte meiosis.Biochim Biophys Acta. 2016 Dec;1863(12):2993-3000. doi: 10.1016/j.bbamcr.2016.09.020. Biochim Biophys Acta. 2016. PMID: 27693251
-
Spindle assembly checkpoint and its regulators in meiosis.Hum Reprod Update. 2012 Jan-Feb;18(1):60-72. doi: 10.1093/humupd/dmr044. Epub 2011 Nov 14. Hum Reprod Update. 2012. PMID: 22086113 Review.
Cited by
-
Gross Chromosomal Rearrangement at Centromeres.Biomolecules. 2023 Dec 24;14(1):28. doi: 10.3390/biom14010028. Biomolecules. 2023. PMID: 38254628 Free PMC article. Review.
-
CDK12 phosphorylates 4E-BP1 to enable mTORC1-dependent translation and mitotic genome stability.Genes Dev. 2019 Apr 1;33(7-8):418-435. doi: 10.1101/gad.322339.118. Epub 2019 Feb 28. Genes Dev. 2019. PMID: 30819820 Free PMC article.
-
Antagonistic control of Caenorhabditis elegans germline stem cell proliferation and differentiation by PUF proteins FBF-1 and FBF-2.Elife. 2020 Aug 17;9:e52788. doi: 10.7554/eLife.52788. Elife. 2020. PMID: 32804074 Free PMC article.
-
Aurora B prevents aneuploidy via MAD2 during the first mitotic cleavage in oxidatively damaged embryos.Cell Prolif. 2019 Sep;52(5):e12657. doi: 10.1111/cpr.12657. Epub 2019 Jul 1. Cell Prolif. 2019. PMID: 31264311 Free PMC article.
-
HPV16-E2 induces prophase arrest and activates the cellular DNA damage response in vitro and in precursor lesions of cervical carcinoma.Oncotarget. 2015 Oct 27;6(33):34979-91. doi: 10.18632/oncotarget.5512. Oncotarget. 2015. PMID: 26474276 Free PMC article.
References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials
Miscellaneous

