Loss of GM130 in breast cancer cells and its effects on cell migration, invasion and polarity
- PMID: 25892554
- PMCID: PMC4612654
- DOI: 10.1080/15384101.2015.1007771
Loss of GM130 in breast cancer cells and its effects on cell migration, invasion and polarity
Abstract
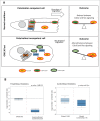
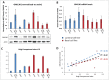
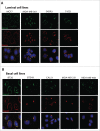
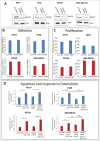
Spatially distinct pools of the small GTPase Cdc42 were observed, but the major focus of research so far has been to investigate its signaling at the plasma membrane. We recently showed that the Golgi pool of Cdc42 is relevant for cell polarity and that it is regulated by GM130, a Golgi matrix protein. Loss of GM130 abrogated cell polarity and consistent with the notion that polarity is frequently impaired in cancer, we found that GM130 is downregulated in colorectal cancer. Whether the loss of GM130 solely affects polarity, or whether it affects other processes relevant for tumorigenesis remains unclear. In a panel of breast cancer cells lines, we investigated the consequences of GM130 depletion on traits of relevance for tumor progression, such as survival, proliferation, adhesion, migration and invasion. We show that cellular assays that depend on polarity, such as chemotaxis and wound scratch assays, are only of limited use to investigate the role of polarity modulators in cancer. Depletion of GM130 increases cellular velocity and increases the invasiveness of breast cancer cells, therefore supporting the view that alterations of polarity contribute to tumor progression.
Figures
Similar articles
-
Spatial control of Cdc42 signalling by a GM130-RasGRF complex regulates polarity and tumorigenesis.Nat Commun. 2014 Sep 11;5:4839. doi: 10.1038/ncomms5839. Nat Commun. 2014. PMID: 25208761 Free PMC article.
-
Endomembrane control of cell polarity: Relevance to cancer.Small GTPases. 2015;6(2):104-7. doi: 10.1080/21541248.2015.1018402. Small GTPases. 2015. PMID: 26156751 Free PMC article. Review.
-
GM130-dependent control of Cdc42 activity at the Golgi regulates centrosome organization.Mol Biol Cell. 2009 Feb;20(4):1192-200. doi: 10.1091/mbc.e08-08-0834. Epub 2008 Dec 24. Mol Biol Cell. 2009. PMID: 19109421 Free PMC article.
-
GM130 regulates epithelial-to-mesenchymal transition and invasion of gastric cancer cells via snail.Int J Clin Exp Pathol. 2015 Sep 1;8(9):10784-91. eCollection 2015. Int J Clin Exp Pathol. 2015. PMID: 26617790 Free PMC article.
-
Emerging new roles of GM130, a cis-Golgi matrix protein, in higher order cell functions.J Pharmacol Sci. 2010;112(3):255-64. doi: 10.1254/jphs.09r03cr. Epub 2010 Mar 2. J Pharmacol Sci. 2010. PMID: 20197635 Review.
Cited by
-
Targeted protein unfolding uncovers a Golgi-specific transcriptional stress response.Mol Biol Cell. 2018 Jun 1;29(11):1284-1298. doi: 10.1091/mbc.E17-11-0693. Epub 2018 Apr 5. Mol Biol Cell. 2018. PMID: 29851555 Free PMC article.
-
Golgi ribbon disassembly during mitosis, differentiation and disease progression.Curr Opin Cell Biol. 2017 Aug;47:43-51. doi: 10.1016/j.ceb.2017.03.008. Epub 2017 Apr 5. Curr Opin Cell Biol. 2017. PMID: 28390244 Free PMC article. Review.
-
The pseudophosphatase STYX targets the F-box of FBXW7 and inhibits SCFFBXW7 function.EMBO J. 2017 Feb 1;36(3):260-273. doi: 10.15252/embj.201694795. Epub 2016 Dec 22. EMBO J. 2017. PMID: 28007894 Free PMC article.
-
Loss of TTC17 promotes breast cancer metastasis through RAP1/CDC42 signaling and sensitizes it to rapamycin and paclitaxel.Cell Biosci. 2023 Mar 9;13(1):50. doi: 10.1186/s13578-023-01004-8. Cell Biosci. 2023. PMID: 36895029 Free PMC article.
-
Intuitive repositioning of an anti-depressant drug in combination with tivozanib: precision medicine for breast cancer therapy.Mol Cell Biochem. 2021 Nov;476(11):4177-4189. doi: 10.1007/s11010-021-04230-1. Epub 2021 Jul 29. Mol Cell Biochem. 2021. PMID: 34324118
References
-
- Ridley AJ. Rho GTPases and actin dynamics in membrane protrusions and vesicle trafficking. Trends in Cell Biology 2006; 16:522-9. - PubMed
-
- Hall A. Rho family GTPases. Biochem Soc Trans 2012; 40:1378-82. - PubMed
-
- Etienne-Manneville S. Cdc42 - the centre of polarity. Journal of Cell Science 2004; 117:1291-300. - PubMed
-
- Wang F, Herzmark P, Weiner OD, Srinivasan S, Servant G, Bourne HR. Lipid products of PI(3)Ks maintain persistent cell polarity and directed motility in neutrophils. Nat Cell Biol 2002; 4:513-8. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Research Materials
Miscellaneous
