Improved survival among colon cancer patients with increased differentially expressed pathways
- PMID: 25890236
- PMCID: PMC4389992
- DOI: 10.1186/s12916-015-0292-9
Improved survival among colon cancer patients with increased differentially expressed pathways
Abstract
Background: Studies of colorectal cancer (CRC) have shown that hundreds to thousands of genes are differentially expressed in tumors when compared to normal tissue samples. In this study, we evaluate how genes that are differentially expressed in colon versus normal tissue influence survival.
Methods: We performed RNA-seq on tumor/normal paired samples from 175 colon cancer patients. We implemented a cross validation strategy to determine genes that were significantly differentially expressed between tumor and normal samples. Differentially expressed genes were evaluated with Ingenuity Pathway Analysis to identify key pathways that were de-regulated. A summary differential pathway expression score (DPES) was developed to summarize hazard of dying while adjusting for age, American Joint Committee on Cancer (AJCC) stage, sex, and tumor molecular phenotype, i.e., MSI, TP53, KRAS, and CIMP.
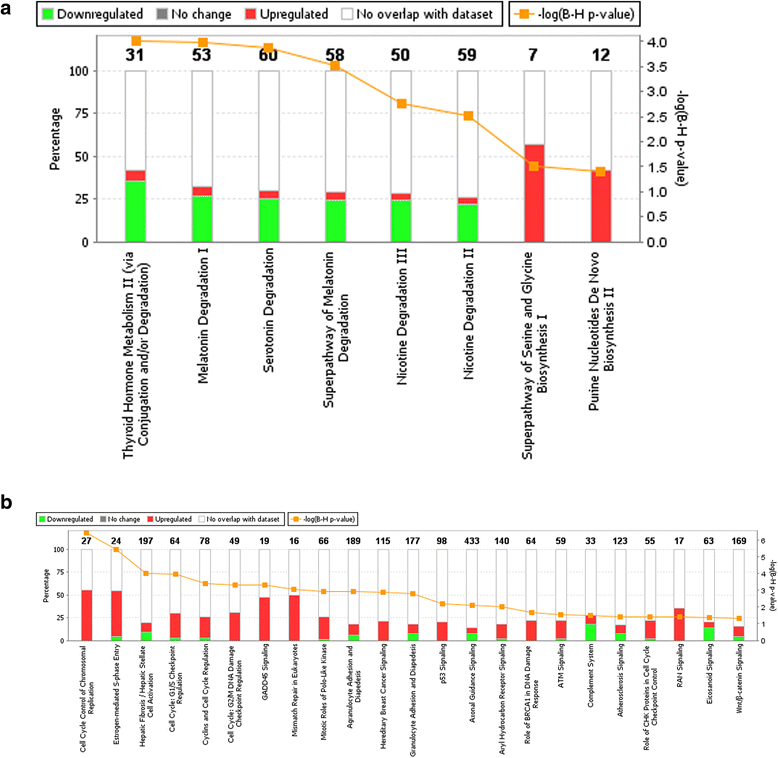
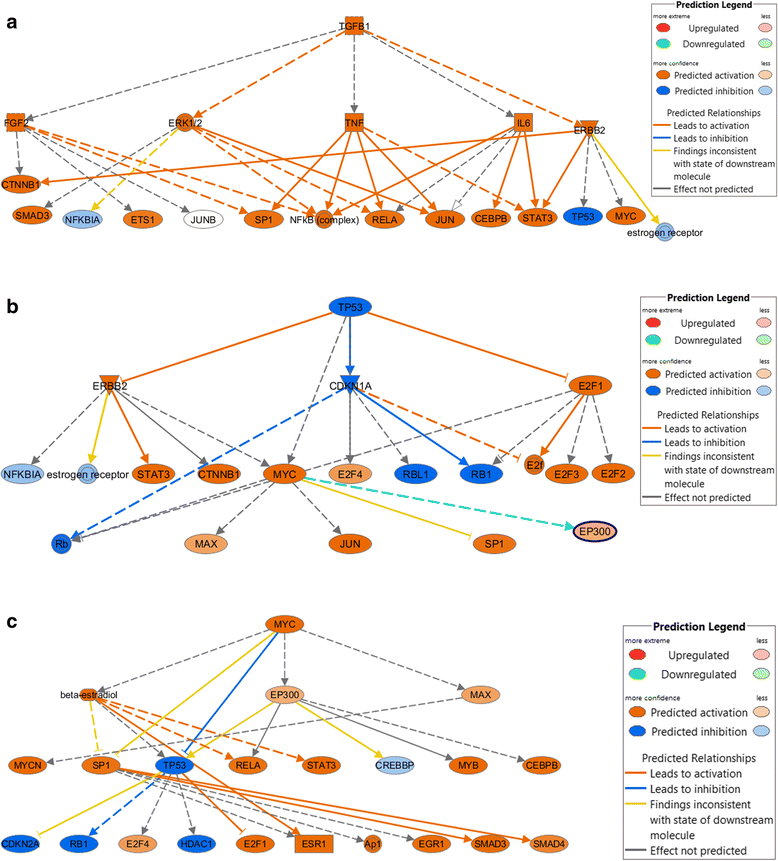
Results: A total of 1,138 genes were up-regulated and 695 were down-regulated. These de-regulated genes were enriched for 19 Ingenuity Canonical Pathways, with the most significant pathways involving cell signaling and growth. Of the enriched pathways, 16 were significantly associated with CRC-specific mortality, including 1 metabolic pathway and 15 signaling pathways. In all instances, having a higher DPES (i.e., more de-regulated genes) was associated with better survival. Further assessment showed that individuals diagnosed at AJCC Stage 1 had more de-regulated genes than individuals diagnosed at AJCC Stage 4.
Conclusions: Our data suggest that having more de-regulated pathways is associated with a good prognosis and may be a reaction to key events that are disabling to tumor progression. Please see related article: http://dx.doi.org/10.1186/s12916-015-0307-6 .
Figures
Comment in
-
Altered pathways and colorectal cancer prognosis.BMC Med. 2015 Apr 8;13:76. doi: 10.1186/s12916-015-0307-6. BMC Med. 2015. PMID: 25885526 Free PMC article.
Similar articles
-
Altered pathways and colorectal cancer prognosis.BMC Med. 2015 Apr 8;13:76. doi: 10.1186/s12916-015-0307-6. BMC Med. 2015. PMID: 25885526 Free PMC article.
-
Gene expression in colon cancer: A focus on tumor site and molecular phenotype.Genes Chromosomes Cancer. 2015 Sep;54(9):527-41. doi: 10.1002/gcc.22265. Epub 2015 Jul 14. Genes Chromosomes Cancer. 2015. PMID: 26171582 Free PMC article.
-
DNA copy number alterations, gene expression changes and disease-free survival in patients with colorectal cancer: a 10 year follow-up.Cell Oncol (Dordr). 2016 Dec;39(6):545-558. doi: 10.1007/s13402-016-0299-z. Epub 2016 Oct 5. Cell Oncol (Dordr). 2016. PMID: 27709558
-
Identification of key pathways and genes in colorectal cancer using bioinformatics analysis.Med Oncol. 2016 Oct;33(10):111. doi: 10.1007/s12032-016-0829-6. Epub 2016 Aug 31. Med Oncol. 2016. PMID: 27581154
-
Molecular biomarkers and classification models in the evaluation of the prognosis of colorectal cancer.Anticancer Res. 2014 May;34(5):2061-8. Anticancer Res. 2014. PMID: 24778007 Review.
Cited by
-
Infrequently expressed miRNAs in colorectal cancer tissue and tumor molecular phenotype.Mod Pathol. 2017 Aug;30(8):1152-1169. doi: 10.1038/modpathol.2017.38. Epub 2017 May 26. Mod Pathol. 2017. PMID: 28548123 Free PMC article.
-
Next-generation sequencing: recent applications to the analysis of colorectal cancer.J Transl Med. 2017 Dec 8;15(1):246. doi: 10.1186/s12967-017-1353-y. J Transl Med. 2017. PMID: 29221448 Free PMC article. Review.
-
Altered pathways and colorectal cancer prognosis.BMC Med. 2015 Apr 8;13:76. doi: 10.1186/s12916-015-0307-6. BMC Med. 2015. PMID: 25885526 Free PMC article.
-
Transcriptome profiling of the rat retina after optic nerve transection.Sci Rep. 2016 Jun 29;6:28736. doi: 10.1038/srep28736. Sci Rep. 2016. PMID: 27353354 Free PMC article.
-
Dietary intake alters gene expression in colon tissue: possible underlying mechanism for the influence of diet on disease.Pharmacogenet Genomics. 2016 Jun;26(6):294-306. doi: 10.1097/FPC.0000000000000217. Pharmacogenet Genomics. 2016. PMID: 26959716 Free PMC article.
References
-
- Birkenkamp-Demtroder K, Christensen LL, Olesen SH, Frederiksen CM, Laiho P, Aaltonen LA, et al. Gene expression in colorectal cancer. Cancer Res. 2002;62:4352–63. - PubMed
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Research Materials
Miscellaneous

