Hedgehog Acyltransferase as a target in estrogen receptor positive, HER2 amplified, and tamoxifen resistant breast cancer cells
- PMID: 25889650
- PMCID: PMC4711017
- DOI: 10.1186/s12943-015-0345-x
Hedgehog Acyltransferase as a target in estrogen receptor positive, HER2 amplified, and tamoxifen resistant breast cancer cells
Abstract
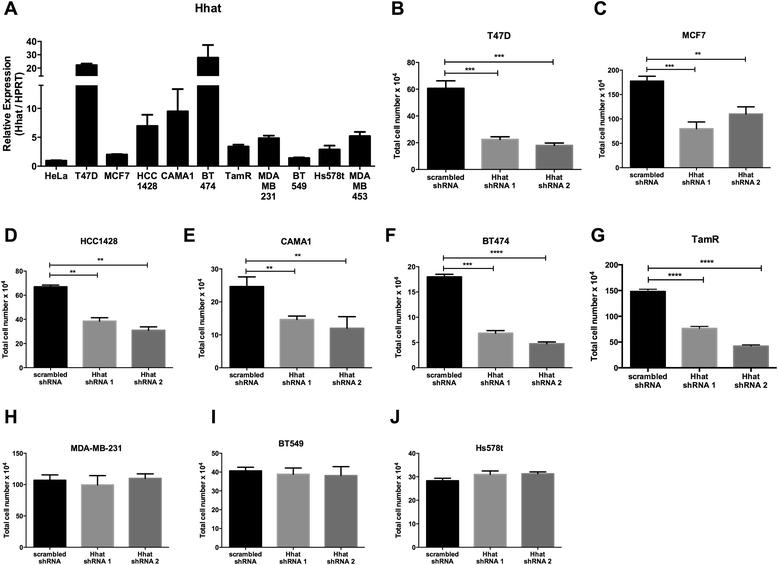
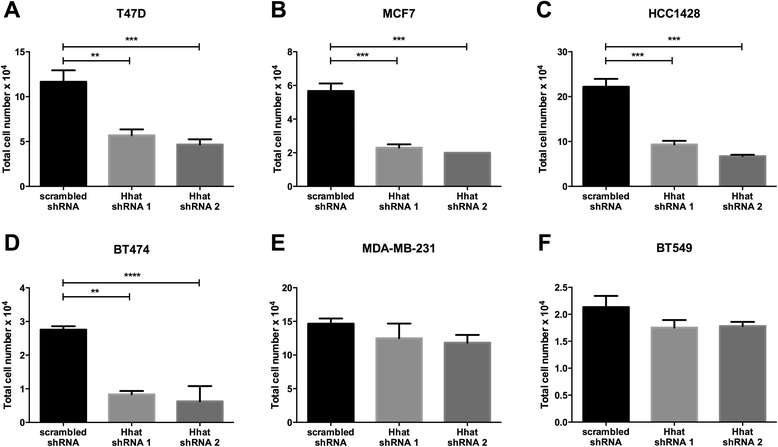
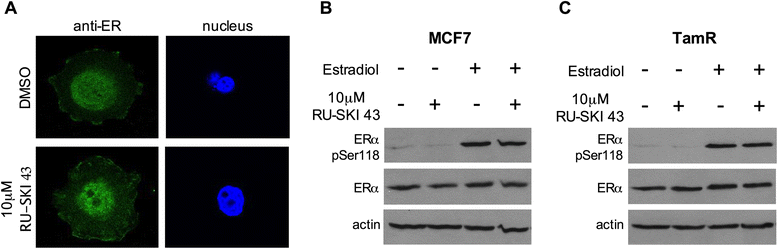
Background: Hedgehog acyltransferase (Hhat) catalyzes the transfer of the fatty acid palmitate onto Sonic Hedgehog (Shh), a modification that is essential for Shh signaling activity. The Shh signaling pathway has been implicated in the progression of breast cancer.
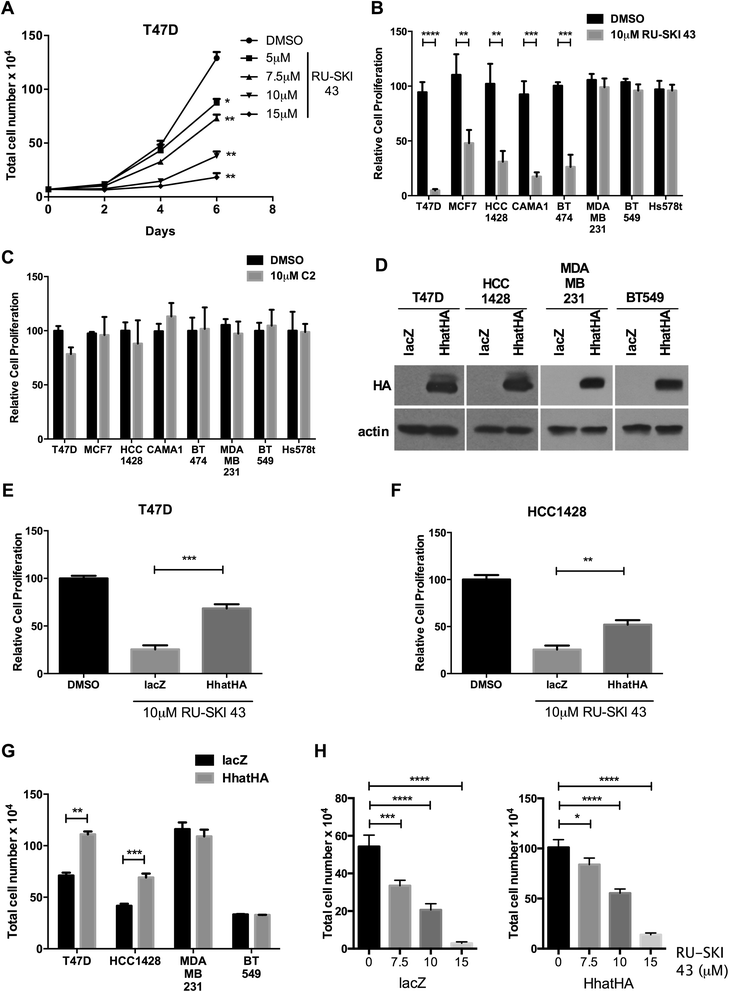
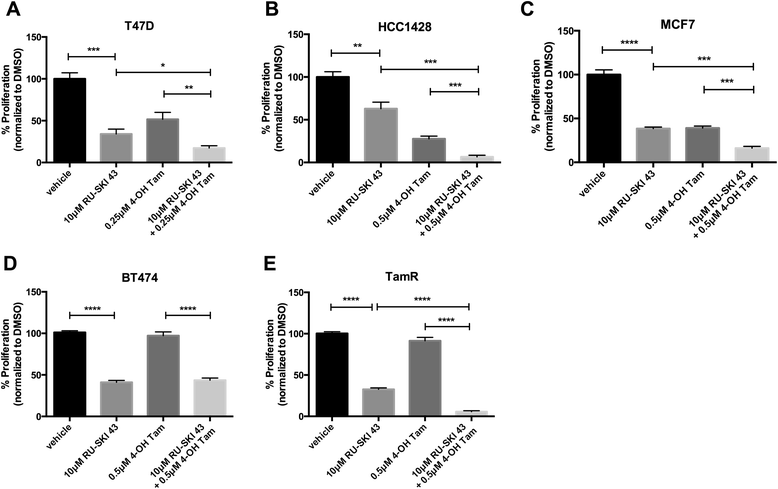
Methods: To determine the functional significance of Hhat expression in breast cancer, we used a panel of breast cancer cell lines that included estrogen receptor (ER) positive, HER2 amplified, triple negative, and tamoxifen resistant cells. We monitored both anchorage dependent and independent proliferation of these cells following depletion of Hhat with lentiviral shRNA and inhibition of Hhat activity with RU-SKI 43, a small molecule inhibitor of Hhat.
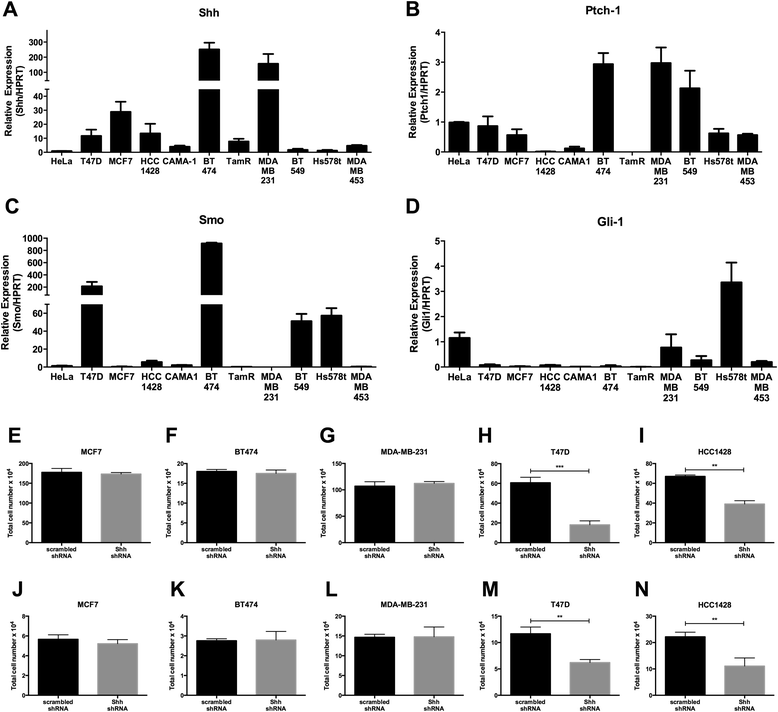
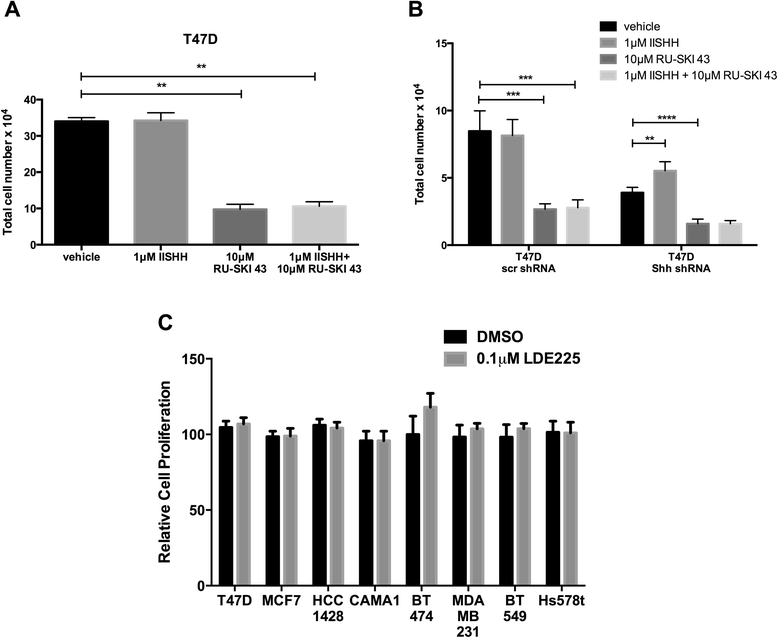
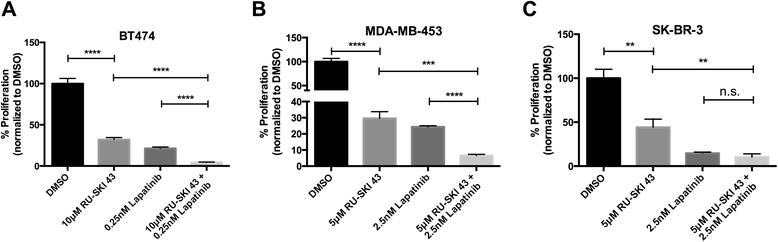
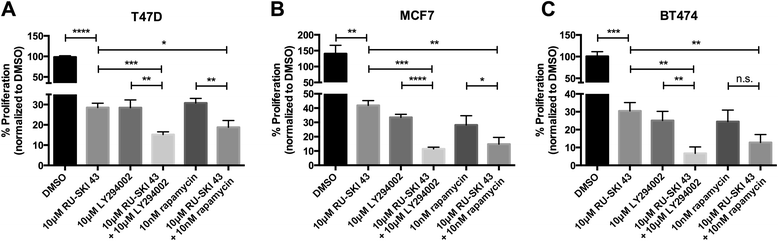
Results: Depletion of Hhat decreased anchorage-dependent and anchorage-independent proliferation of ER positive, but not triple negative, breast cancer cells. Treatment with RU-SKI 43 also reduced ER positive cell proliferation, whereas a structurally related, inactive compound had no effect. Overexpression of Hhat in ER positive cells not only rescued the growth defect in the presence of RU-SKI 43 but also resulted in increased cell proliferation in the absence of drug. Furthermore, depletion or inhibition of Hhat reduced proliferation of HER2 amplified as well as tamoxifen resistant cells. Inhibition of Smoothened had no effect on proliferation, indicating that canonical Shh signaling was not operative. Moreover, Hhat regulated the proliferation of both Shh responsive and non-responsive ER positive cells, suggesting a Shh independent function for Hhat.
Conclusions: These data suggest that Hhat plays a critical role in ER positive, HER2 amplified, and hormone resistant breast cancer proliferation and highlights the potential promise of Hhat inhibitors for therapeutic benefit in breast cancer.
Figures
Similar articles
-
Hedgehog acyltransferase as a target in pancreatic ductal adenocarcinoma.Oncogene. 2015 Jan 8;34(2):263-8. doi: 10.1038/onc.2013.575. Epub 2014 Jan 27. Oncogene. 2015. PMID: 24469057 Free PMC article.
-
Mechanisms of tamoxifen resistance: increased estrogen receptor-HER2/neu cross-talk in ER/HER2-positive breast cancer.J Natl Cancer Inst. 2004 Jun 16;96(12):926-35. doi: 10.1093/jnci/djh166. J Natl Cancer Inst. 2004. PMID: 15199112
-
Dacomitinib (PF-00299804), an irreversible Pan-HER inhibitor, inhibits proliferation of HER2-amplified breast cancer cell lines resistant to trastuzumab and lapatinib.Mol Cancer Ther. 2012 Sep;11(9):1978-87. doi: 10.1158/1535-7163.MCT-11-0730. Epub 2012 Jul 3. Mol Cancer Ther. 2012. PMID: 22761403
-
Palmitoylation of Hedgehog proteins by Hedgehog acyltransferase: roles in signalling and disease.Open Biol. 2021 Mar;11(3):200414. doi: 10.1098/rsob.200414. Epub 2021 Mar 3. Open Biol. 2021. PMID: 33653085 Free PMC article. Review.
-
Inhibition of erbB receptor (HER) tyrosine kinases as a strategy to abrogate antiestrogen resistance in human breast cancer.Clin Cancer Res. 2001 Dec;7(12 Suppl):4436s-4442s; discussion 4411s-4412s. Clin Cancer Res. 2001. PMID: 11916237 Review.
Cited by
-
PRKCI Mediates Radiosensitivity via the Hedgehog/GLI1 Pathway in Cervical Cancer.Front Oncol. 2022 Jun 16;12:887139. doi: 10.3389/fonc.2022.887139. eCollection 2022. Front Oncol. 2022. PMID: 35785194 Free PMC article.
-
Targeting the Sonic Hedgehog Signaling Pathway: Review of Smoothened and GLI Inhibitors.Cancers (Basel). 2016 Feb 15;8(2):22. doi: 10.3390/cancers8020022. Cancers (Basel). 2016. PMID: 26891329 Free PMC article. Review.
-
The role of Hedgehog and Notch signaling pathway in cancer.Mol Biomed. 2022 Dec 15;3(1):44. doi: 10.1186/s43556-022-00099-8. Mol Biomed. 2022. PMID: 36517618 Free PMC article. Review.
-
NKX6-1 mediates cancer stem-like properties and regulates sonic hedgehog signaling in leiomyosarcoma.J Biomed Sci. 2021 Apr 27;28(1):32. doi: 10.1186/s12929-021-00726-6. J Biomed Sci. 2021. PMID: 33906647 Free PMC article.
-
Design, Synthesis, and Evaluation of Inhibitors of Hedgehog Acyltransferase.J Med Chem. 2024 Jan 25;67(2):1061-1078. doi: 10.1021/acs.jmedchem.3c01363. Epub 2024 Jan 10. J Med Chem. 2024. PMID: 38198226 Free PMC article.
References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Research Materials
Miscellaneous

