Transcriptome analysis of ageing in uninjured human Achilles tendon
- PMID: 25888722
- PMCID: PMC4355574
- DOI: 10.1186/s13075-015-0544-2
Transcriptome analysis of ageing in uninjured human Achilles tendon
Abstract
Introduction: The risk of tendon injury and disease increases significantly with increasing age. The aim of the study was to characterise transcriptional changes in human Achilles tendon during the ageing process in order to identify molecular signatures that might contribute to age-related degeneration.
Methods: RNA for gene expression analysis using RNA-Seq and quantitative real-time polymerase chain reaction analysis was isolated from young and old macroscopically normal human Achilles tendon. RNA sequence libraries were prepared following ribosomal RNA depletion, and sequencing was undertaken by using the Illumina HiSeq 2000 platform. Expression levels among genes were compared by using fragments per kilobase of exon per million fragments mapped. Differentially expressed genes were defined by using Benjamini-Hochberg false discovery rate approach (P<0.05, expression ratios 1.4 log2 fold change). Alternative splicing of exon variants were also examined by using Cufflinks. The functional significance of genes that showed differential expression between young and old tendon was determined by using ingenuity pathway analysis.
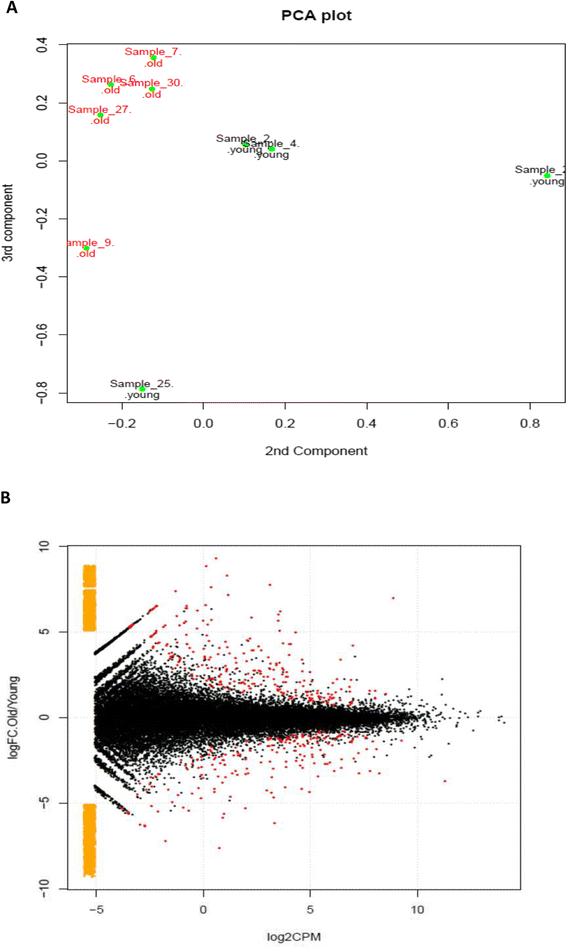
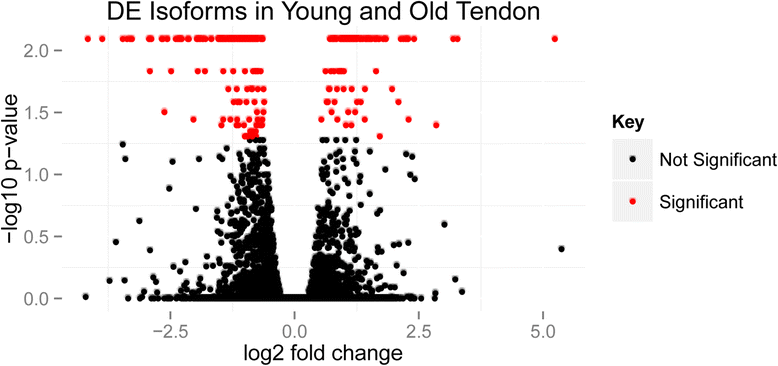
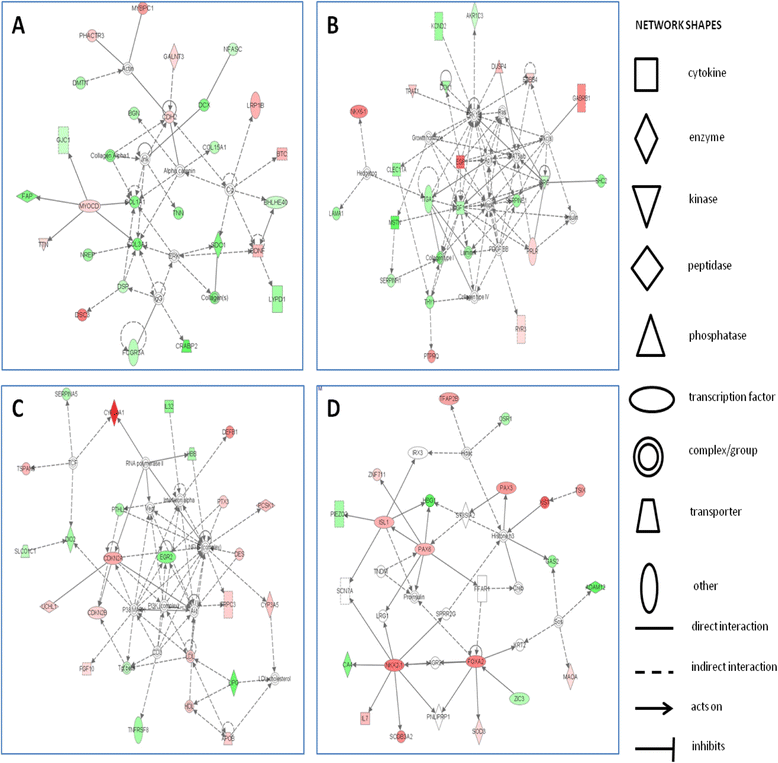
Results: In total, the expression of 325 transcribed elements, including protein-coding transcripts and non-coding transcripts (small non-coding RNAs, pseudogenes, long non-coding RNAs and a single microRNA), was significantly different in old compared with young tendon (±1.4 log2 fold change, P<0.05). Of these, 191 were at higher levels in older tendon and 134 were at lower levels in older tendon. The top networks for genes differentially expressed with tendon age were from cellular function, cellular growth, and cellular cycling pathways. Notable differential transcriptome changes were also observed in alternative splicing patterns. Several of the top gene ontology terms identified in downregulated isoforms in old tendon related to collagen and post-translational modification of collagen.
Conclusions: This study demonstrates dynamic alterations in RNA with age at numerous genomic levels, indicating changes in the regulation of transcriptional networks. The results suggest that ageing is not primarily associated with loss of ability to synthesise matrix proteins and matrix-degrading enzymes. In addition, we have identified non-coding RNA genes and differentially expressed transcript isoforms of known matrix components with ageing which require further investigation.
Figures
Similar articles
-
Transcriptomic signatures in cartilage ageing.Arthritis Res Ther. 2013 Aug 23;15(4):R98. doi: 10.1186/ar4278. Arthritis Res Ther. 2013. PMID: 23971731 Free PMC article.
-
Dynamics of the transcriptional landscape during human fetal testis and ovary development.Hum Reprod. 2020 May 1;35(5):1099-1119. doi: 10.1093/humrep/deaa041. Hum Reprod. 2020. PMID: 32412604
-
Differing molecular response of young and advanced maternal age human oocytes to IVM.Hum Reprod. 2017 Nov 1;32(11):2199-2208. doi: 10.1093/humrep/dex284. Hum Reprod. 2017. PMID: 29025019 Free PMC article.
-
Whole transcriptome sequencing of the aging rat brain reveals dynamic RNA changes in the dark matter of the genome.Age (Dordr). 2013 Jun;35(3):763-76. doi: 10.1007/s11357-012-9410-1. Epub 2012 May 4. Age (Dordr). 2013. PMID: 22555619 Free PMC article.
-
Comparative transcriptome analysis of epithelial and fiber cells in newborn mouse lenses with RNA sequencing.Mol Vis. 2014 Nov 4;20:1491-517. eCollection 2014. Mol Vis. 2014. PMID: 25489224 Free PMC article.
Cited by
-
Structure-function specialisation of the interfascicular matrix in the human achilles tendon.Acta Biomater. 2021 Sep 1;131:381-390. doi: 10.1016/j.actbio.2021.07.019. Epub 2021 Jul 13. Acta Biomater. 2021. PMID: 34271169 Free PMC article.
-
Serum snoRNAs as biomarkers for joint ageing and post traumatic osteoarthritis.Sci Rep. 2017 Mar 2;7:43558. doi: 10.1038/srep43558. Sci Rep. 2017. PMID: 28252005 Free PMC article.
-
Minimally invasive, endoscopic Achilles tendon reconstruction using semitendinosus and gracilis tendons with Endobutton stabilization.BMC Musculoskelet Disord. 2016 Jun 3;17:247. doi: 10.1186/s12891-016-1099-3. BMC Musculoskelet Disord. 2016. PMID: 27256340 Free PMC article.
-
Age-related changes in microRNAs expression in cruciate ligaments of wild-stock house mice.Physiol Rep. 2022 Aug;10(16):e15426. doi: 10.14814/phy2.15426. Physiol Rep. 2022. PMID: 35993414 Free PMC article.
-
Pleiotropic roles of the matricellular protein Sparc in tendon maturation and ageing.Sci Rep. 2016 Sep 2;6:32635. doi: 10.1038/srep32635. Sci Rep. 2016. PMID: 27586416 Free PMC article.
References
-
- Beard JR, Biggs S, Bloom BD, Fried LP, Hogan L, Kalache A, et al. Global Population Ageing: Peril or Promise. Geneva: World Economic Forum; 2011.
-
- Corps AN, Robinson AH, Harrall RL, Avery NC, Curry VA, Hazleman BL, et al. Changes in matrix protein biochemistry and the expression of mRNA encoding matrix proteins and metalloproteinases in posterior tibialis tendinopathy. Ann Rheum Dis. 2012;71:746–52. doi: 10.1136/annrheumdis-2011-200391. - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases

