ALDsuite: Dense marker MALD using principal components of ancestral linkage disequilibrium
- PMID: 25886794
- PMCID: PMC4408589
- DOI: 10.1186/s12863-015-0179-y
ALDsuite: Dense marker MALD using principal components of ancestral linkage disequilibrium
Abstract
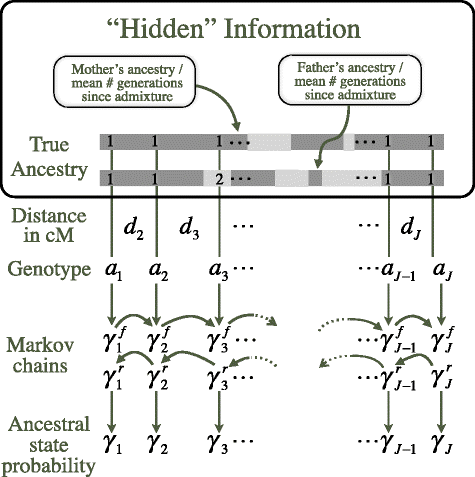
Background: Mapping by admixture linkage disequilibrium (MALD) is a whole genome gene mapping method that uses LD from extended blocks of ancestry inherited from parental populations among admixed individuals to map associations for diseases, that vary in prevalence among human populations. The extended LD queried for marker association with ancestry results in a greatly reduced number of comparisons compared to standard genome wide association studies. As ancestral population LD tends to confound the analysis of admixture LD, the earliest algorithms for MALD required marker sets sufficiently sparse to lack significant ancestral LD between markers. However current genotyping technologies routinely provide dense SNP data, which convey more information than sparse sets, if this information can be efficiently used. There are currently no software solutions that offer both local ancestry inference using dense marker data and disease association statistics.
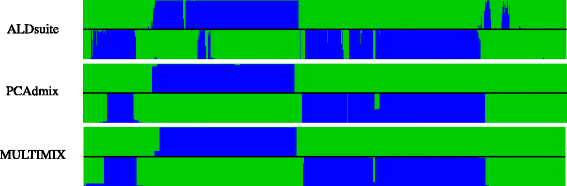
Results: We present here an R package, ALDsuite, which accounts for local LD using principal components of haplotypes from surrogate ancestral population data, and includes tools for quality control of data, MALD, downstream analysis of results and visualization graphics.
Conclusions: ALDsuite offers a fast, accurate estimation of global and local ancestry and comes bundled with the tools needed for MALD, from data quality control through mapping of and visualization of disease genes.
Figures
Similar articles
-
Mapping asthma-associated variants in admixed populations.Front Genet. 2015 Sep 29;6:292. doi: 10.3389/fgene.2015.00292. eCollection 2015. Front Genet. 2015. PMID: 26483834 Free PMC article. Review.
-
Inferring ancestries efficiently in admixed populations with linkage disequilibrium.J Comput Biol. 2009 Aug;16(8):1141-50. doi: 10.1089/cmb.2009.0105. J Comput Biol. 2009. PMID: 19645595
-
The Analysis of Ethnic Mixtures.Methods Mol Biol. 2017;1666:505-525. doi: 10.1007/978-1-4939-7274-6_25. Methods Mol Biol. 2017. PMID: 28980262
-
The analysis of ethnic mixtures.Methods Mol Biol. 2012;850:465-81. doi: 10.1007/978-1-61779-555-8_25. Methods Mol Biol. 2012. PMID: 22307714 Free PMC article.
-
Mapping by admixture linkage disequilibrium: advances, limitations and guidelines.Nat Rev Genet. 2005 Aug;6(8):623-32. doi: 10.1038/nrg1657. Nat Rev Genet. 2005. PMID: 16012528 Review.
Cited by
-
Including diverse and admixed populations in genetic epidemiology research.Genet Epidemiol. 2022 Oct;46(7):347-371. doi: 10.1002/gepi.22492. Epub 2022 Jul 16. Genet Epidemiol. 2022. PMID: 35842778 Free PMC article.
-
Mapping asthma-associated variants in admixed populations.Front Genet. 2015 Sep 29;6:292. doi: 10.3389/fgene.2015.00292. eCollection 2015. Front Genet. 2015. PMID: 26483834 Free PMC article. Review.
References
-
- Nalls MA, Wilson JG, Patterson NJ, Tandon A, Zmuda JM, Huntsman S, et al. Admixture mapping of white cell count: genetic locus responsible for lower white blood cell count in the Health ABC and Jackson Heart studies. Am J Human Genet. 2008;82(1):81–7. doi: 10.1016/j.ajhg.2007.09.003. - DOI - PMC - PubMed
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials

