ANDSystem: an Associative Network Discovery System for automated literature mining in the field of biology
- PMID: 25881313
- PMCID: PMC4407185
- DOI: 10.1186/1752-0509-9-S2-S2
ANDSystem: an Associative Network Discovery System for automated literature mining in the field of biology
Abstract
Background: Sufficient knowledge of molecular and genetic interactions, which comprise the entire basis of the functioning of living systems, is one of the necessary requirements for successfully answering almost any research question in the field of biology and medicine. To date, more than 24 million scientific papers can be found in PubMed, with many of them containing descriptions of a wide range of biological processes. The analysis of such tremendous amounts of data requires the use of automated text-mining approaches. Although a handful of tools have recently been developed to meet this need, none of them provide error-free extraction of highly detailed information.
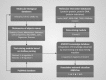
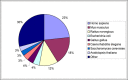
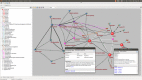
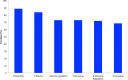
Results: The ANDSystem package was developed for the reconstruction and analysis of molecular genetic networks based on an automated text-mining technique. It provides a detailed description of the various types of interactions between genes, proteins, microRNA's, metabolites, cellular components, pathways and diseases, taking into account the specificity of cell lines and organisms. Although the accuracy of ANDSystem is comparable to other well known text-mining tools, such as Pathway Studio and STRING, it outperforms them in having the ability to identify an increased number of interaction types.
Conclusion: The use of ANDSystem, in combination with Pathway Studio and STRING, can improve the quality of the automated reconstruction of molecular and genetic networks. ANDSystem should provide a useful tool for researchers working in a number of different fields, including biology, biotechnology, pharmacology and medicine.
Figures



Similar articles
-
ANDVisio: a new tool for graphic visualization and analysis of literature mined associative gene networks in the ANDSystem.In Silico Biol. 2011-2012;11(3-4):149-61. doi: 10.3233/ISB-2012-0449. In Silico Biol. 2011. PMID: 22935968
-
A new version of the ANDSystem tool for automatic extraction of knowledge from scientific publications with expanded functionality for reconstruction of associative gene networks by considering tissue-specific gene expression.BMC Bioinformatics. 2019 Feb 5;20(Suppl 1):34. doi: 10.1186/s12859-018-2567-6. BMC Bioinformatics. 2019. PMID: 30717676 Free PMC article.
-
Interactome of the hepatitis C virus: Literature mining with ANDSystem.Virus Res. 2016 Jun 15;218:40-8. doi: 10.1016/j.virusres.2015.12.003. Epub 2015 Dec 7. Virus Res. 2016. PMID: 26673098
-
Mining biological networks from full-text articles.Methods Mol Biol. 2014;1159:135-45. doi: 10.1007/978-1-4939-0709-0_8. Methods Mol Biol. 2014. PMID: 24788265 Review.
-
Text mining for systems biology.Drug Discov Today. 2014 Feb;19(2):140-4. doi: 10.1016/j.drudis.2013.09.012. Epub 2013 Sep 23. Drug Discov Today. 2014. PMID: 24070668 Review.
Cited by
-
Comorbidity of asthma and hypertension may be mediated by shared genetic dysregulation and drug side effects.Sci Rep. 2019 Nov 8;9(1):16302. doi: 10.1038/s41598-019-52762-w. Sci Rep. 2019. PMID: 31705029 Free PMC article.
-
Prioritization of genes involved in endothelial cell apoptosis by their implication in lymphedema using an analysis of associative gene networks with ANDSystem.BMC Med Genomics. 2019 Mar 13;12(Suppl 2):47. doi: 10.1186/s12920-019-0492-9. BMC Med Genomics. 2019. PMID: 30871556 Free PMC article.
-
Research Topics of the Bioinformatics of Gene Regulation.Int J Mol Sci. 2023 May 15;24(10):8774. doi: 10.3390/ijms24108774. Int J Mol Sci. 2023. PMID: 37240120 Free PMC article.
-
A Survey of Gene Prioritization Tools for Mendelian and Complex Human Diseases.J Integr Bioinform. 2019 Sep 9;16(4):20180069. doi: 10.1515/jib-2018-0069. J Integr Bioinform. 2019. PMID: 31494632 Free PMC article. Review.
-
Associative gene networks reveal novel candidates important for ADHD and dyslexia comorbidity.BMC Med Genomics. 2023 Sep 4;16(1):208. doi: 10.1186/s12920-023-01502-1. BMC Med Genomics. 2023. PMID: 37667328 Free PMC article.
References
-
- Nikitin A, Egorov S, Daraselia N, Mazo I. Pathway studio--the analysis and navigation of molecular networks. Bioinformatics. 2003;1;19(16):2155–7. - PubMed
-
- Jenssen TK, Laegreid A, Komorowski J, Hovig E. A literature network of human genes for high-throughput analysis of gene expression. Nat Genet. 2001;28(1):21–8. - PubMed
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources

