Low levels of polymorphisms and no evidence for diversifying selection on the Plasmodium knowlesi Apical Membrane Antigen 1 gene
- PMID: 25881166
- PMCID: PMC4400157
- DOI: 10.1371/journal.pone.0124400
Low levels of polymorphisms and no evidence for diversifying selection on the Plasmodium knowlesi Apical Membrane Antigen 1 gene
Abstract
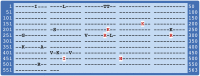
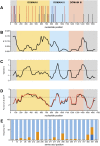
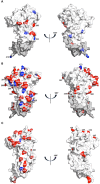
Infection with Plasmodium knowlesi, a zoonotic primate malaria, is a growing human health problem in Southeast Asia. P. knowlesi is being used in malaria vaccine studies, and a number of proteins are being considered as candidate malaria vaccine antigens, including the Apical Membrane Antigen 1 (AMA1). In order to determine genetic diversity of the ama1 gene and to identify epitopes of AMA1 under strongest immune selection, the ama1 gene of 52 P. knowlesi isolates derived from human infections was sequenced. Sequence analysis of isolates from two geographically isolated regions in Sarawak showed that polymorphism in the protein is low compared to that of AMA1 of the major human malaria parasites, P. falciparum and P. vivax. Although the number of haplotypes was 27, the frequency of mutations at the majority of the polymorphic positions was low, and only six positions had a variance frequency higher than 10%. Only two positions had more than one alternative amino acid. Interestingly, three of the high-frequency polymorphic sites correspond to invariant sites in PfAMA1 or PvAMA1. Statistically significant differences in the quantity of three of the six high frequency mutations were observed between the two regions. These analyses suggest that the pkama1 gene is not under balancing selection, as observed for pfama1 and pvama1, and that the PkAMA1 protein is not a primary target for protective humoral immune responses in their reservoir macaque hosts, unlike PfAMA1 and PvAMA1 in humans. The low level of polymorphism justifies the development of a single allele PkAMA1-based vaccine.
Conflict of interest statement
Figures
Similar articles
-
Plasmodium vivax: sequence polymorphism and effect of natural selection at apical membrane antigen 1 (PvAMA1) among Indian population.Gene. 2008 Aug 1;419(1-2):35-42. doi: 10.1016/j.gene.2008.04.012. Epub 2008 Apr 28. Gene. 2008. PMID: 18547744
-
Population genetics, sequence diversity and selection in the gene encoding the Plasmodium falciparum apical membrane antigen 1 in clinical isolates from the south-east of Iran.Infect Genet Evol. 2013 Jul;17:51-61. doi: 10.1016/j.meegid.2013.03.042. Epub 2013 Apr 2. Infect Genet Evol. 2013. PMID: 23557839
-
Analysis of polymorphisms and selective pressures on ama1 gene in Plasmodium knowlesi isolates from Sabah, Malaysia.J Genet. 2017 Sep;96(4):653-663. doi: 10.1007/s12041-017-0817-4. J Genet. 2017. PMID: 28947714
-
Apical membrane antigen 1: a malaria vaccine candidate in review.Trends Parasitol. 2008 Feb;24(2):74-84. doi: 10.1016/j.pt.2007.12.002. Epub 2008 Jan 15. Trends Parasitol. 2008. PMID: 18226584 Review.
-
Natural selection on apical membrane antigen-1 of Plasmodium falciparum.Parassitologia. 1999 Sep;41(1-3):93-5. Parassitologia. 1999. PMID: 10697839 Review.
Cited by
-
Natural selection on apical membrane antigen 1 (AMA1) of an emerging zoonotic malaria parasite Plasmodium inui.Sci Rep. 2024 Oct 9;14(1):23637. doi: 10.1038/s41598-024-74785-8. Sci Rep. 2024. PMID: 39384839 Free PMC article.
-
The Plasmodium knowlesi Pk41 surface protein diversity, natural selection, sub population and geographical clustering: a 6-cysteine protein family member.PeerJ. 2018 Dec 14;6:e6141. doi: 10.7717/peerj.6141. eCollection 2018. PeerJ. 2018. PMID: 30581686 Free PMC article.
-
Is there evidence of sustained human-mosquito-human transmission of the zoonotic malaria Plasmodium knowlesi? A systematic literature review.Malar J. 2022 Mar 17;21(1):89. doi: 10.1186/s12936-022-04110-z. Malar J. 2022. PMID: 35300703 Free PMC article.
-
Structural patterns of selection and diversity for Plasmodium vivax antigens DBP and AMA1.Malar J. 2018 May 2;17(1):183. doi: 10.1186/s12936-018-2324-3. Malar J. 2018. PMID: 29720179 Free PMC article.
-
Clustering and genetic differentiation of the normocyte binding protein (nbpxa) of Plasmodium knowlesi clinical isolates from Peninsular Malaysia and Malaysia Borneo.Malar J. 2016 Apr 26;15:241. doi: 10.1186/s12936-016-1294-6. Malar J. 2016. PMID: 27118390 Free PMC article.
References
-
- Chin W, Contacos PG, Collins WE, Jeter MH, Alpert E (1968) Experimental mosquito-transmission of Plasmodium knowlesi to man and monkey. Am J Trop Med Hyg 17: 355–358. - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
- Actions
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical

