Genomic variation in tomato, from wild ancestors to contemporary breeding accessions
- PMID: 25880392
- PMCID: PMC4404671
- DOI: 10.1186/s12864-015-1444-1
Genomic variation in tomato, from wild ancestors to contemporary breeding accessions
Abstract
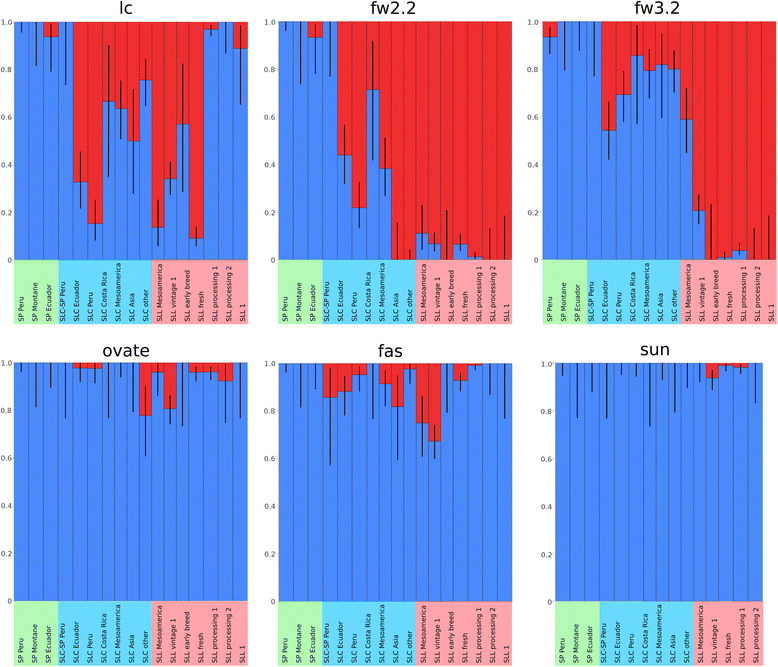
Background: Domestication modifies the genomic variation of species. Quantifying this variation provides insights into the domestication process, facilitates the management of resources used by breeders and germplasm centers, and enables the design of experiments to associate traits with genes. We described and analyzed the genetic diversity of 1,008 tomato accessions including Solanum lycopersicum var. lycopersicum (SLL), S. lycopersicum var. cerasiforme (SLC), and S. pimpinellifolium (SP) that were genotyped using 7,720 SNPs. Additionally, we explored the allelic frequency of six loci affecting fruit weight and shape to infer patterns of selection.
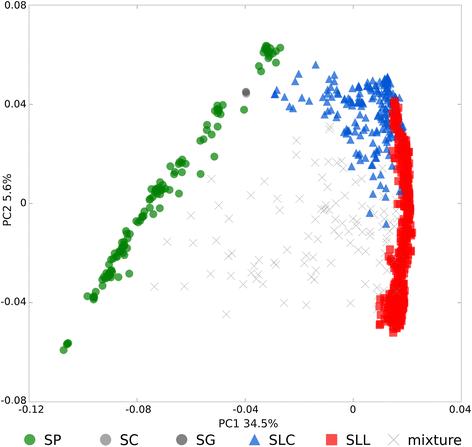
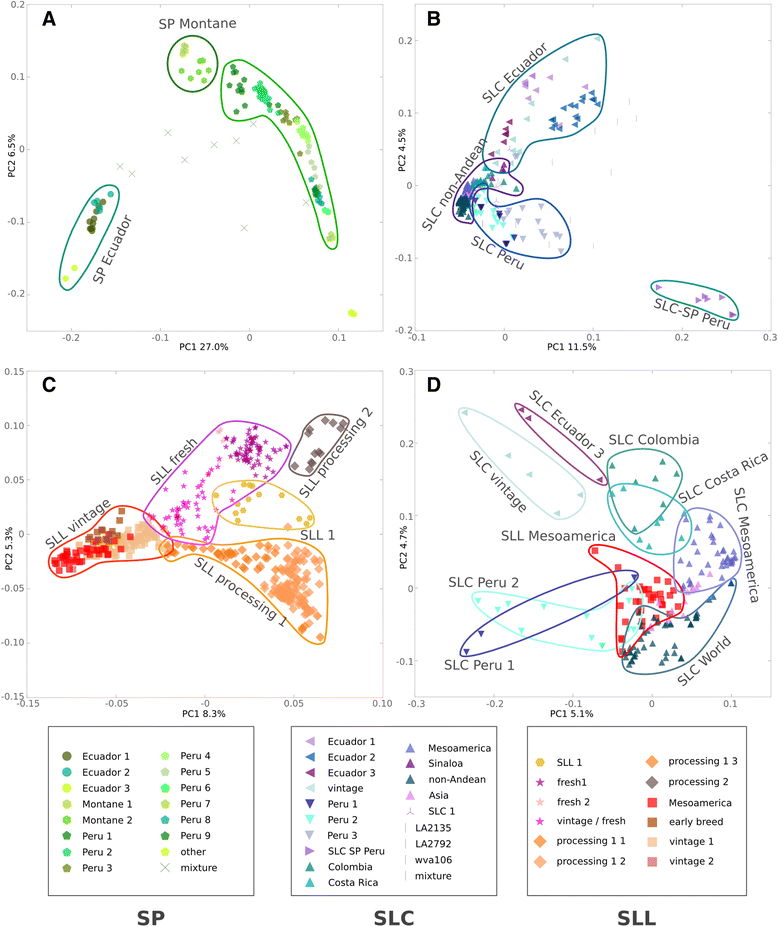
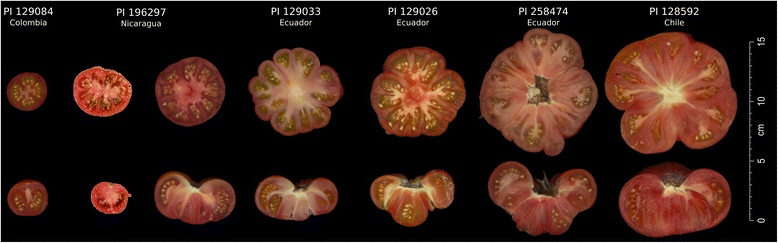
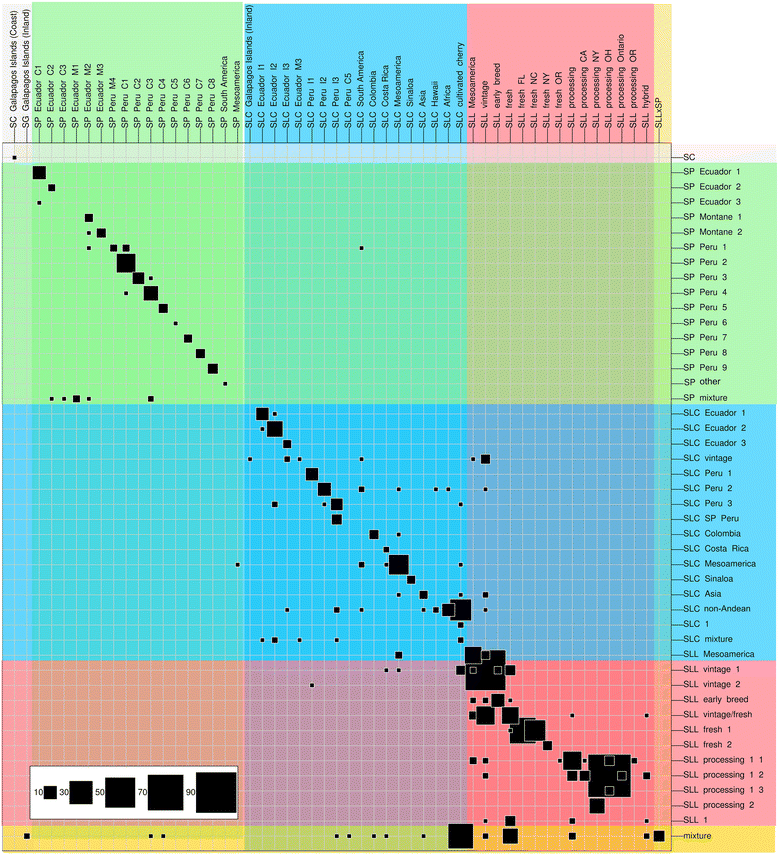
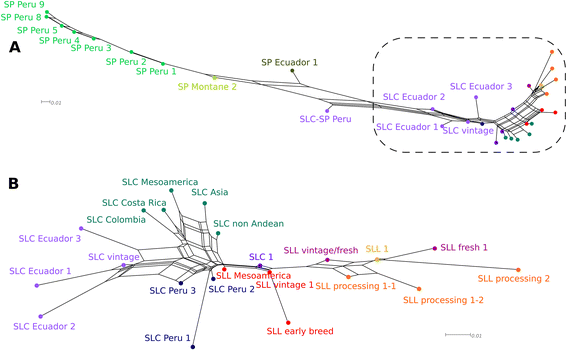
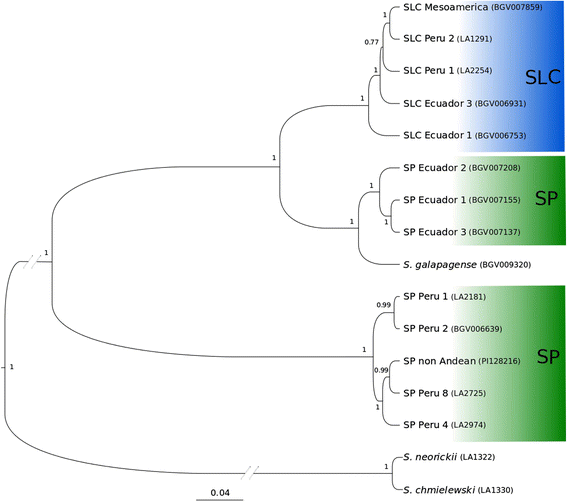
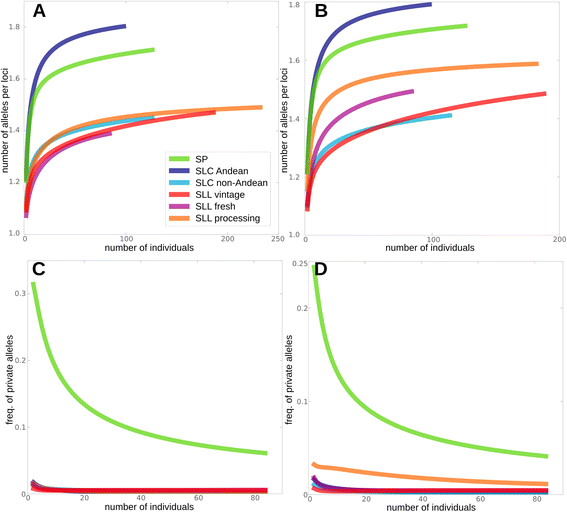
Results: Our results revealed a pattern of variation that strongly supported a two-step domestication process, occasional hybridization in the wild, and differentiation through human selection. These interpretations were consistent with the observed allele frequencies for the six loci affecting fruit weight and shape. Fruit weight was strongly selected in SLC in the Andean region of Ecuador and Northern Peru prior to the domestication of tomato in Mesoamerica. Alleles affecting fruit shape were differentially selected among SLL genetic subgroups. Our results also clarified the biological status of SLC. True SLC was phylogenetically positioned between SP and SLL and its fruit morphology was diverse. SLC and "cherry tomato" are not synonymous terms. The morphologically-based term "cherry tomato" included some SLC, contemporary varieties, as well as many admixtures between SP and SLL. Contemporary SLL showed a moderate increase in nucleotide diversity, when compared with vintage groups.
Conclusions: This study presents a broad and detailed representation of the genomic variation in tomato. Tomato domestication seems to have followed a two step-process; a first domestication in South America and a second step in Mesoamerica. The distribution of fruit weight and shape alleles supports that domestication of SLC occurred in the Andean region. Our results also clarify the biological status of SLC as true phylogenetic group within tomato. We detect Ecuadorian and Peruvian accessions that may represent a pool of unexplored variation that could be of interest for crop improvement.
Figures
Similar articles
-
Variation revealed by SNP genotyping and morphology provides insight into the origin of the tomato.PLoS One. 2012;7(10):e48198. doi: 10.1371/journal.pone.0048198. Epub 2012 Oct 31. PLoS One. 2012. PMID: 23118951 Free PMC article.
-
Genomic Evidence for Complex Domestication History of the Cultivated Tomato in Latin America.Mol Biol Evol. 2020 Apr 1;37(4):1118-1132. doi: 10.1093/molbev/msz297. Mol Biol Evol. 2020. PMID: 31912142 Free PMC article.
-
Exploration of a Resequenced Tomato Core Collection for Phenotypic and Genotypic Variation in Plant Growth and Fruit Quality Traits.Genes (Basel). 2020 Oct 29;11(11):1278. doi: 10.3390/genes11111278. Genes (Basel). 2020. PMID: 33137951 Free PMC article.
-
Domestication and breeding of tomatoes: what have we gained and what can we gain in the future?Ann Bot. 2007 Nov;100(5):1085-94. doi: 10.1093/aob/mcm150. Epub 2007 Aug 23. Ann Bot. 2007. PMID: 17717024 Free PMC article. Review.
-
Genome editing as a tool to achieve the crop ideotype and de novo domestication of wild relatives: Case study in tomato.Plant Sci. 2017 Mar;256:120-130. doi: 10.1016/j.plantsci.2016.12.012. Epub 2016 Dec 28. Plant Sci. 2017. PMID: 28167025 Review.
Cited by
-
Food security and the cultural heritage missing link.Glob Food Sec. 2022 Dec;35:100660. doi: 10.1016/j.gfs.2022.100660. Glob Food Sec. 2022. PMID: 36483217 Free PMC article.
-
Selected genome regions for fruit weight and shelf life in tomato RILs discernible by markers based on genomic sequence information.Breed Sci. 2019 Sep;69(3):447-454. doi: 10.1270/jsbbs.19015. Epub 2019 Jul 4. Breed Sci. 2019. PMID: 31598077 Free PMC article.
-
Flavor and Other Quality Traits of Tomato Cultivars Bred for Diverse Production Systems as Revealed in Organic Low-Input Management.Front Nutr. 2022 Jul 14;9:916642. doi: 10.3389/fnut.2022.916642. eCollection 2022. Front Nutr. 2022. PMID: 35911109 Free PMC article.
-
Breeding Has Increased the Diversity of Cultivated Tomato in The Netherlands.Front Plant Sci. 2019 Dec 20;10:1606. doi: 10.3389/fpls.2019.01606. eCollection 2019. Front Plant Sci. 2019. PMID: 31921253 Free PMC article.
-
Genomic basis of selective breeding from the closest wild relative of large-fruited tomato.Hortic Res. 2023 Jul 8;10(8):uhad142. doi: 10.1093/hr/uhad142. eCollection 2023 Aug. Hortic Res. 2023. PMID: 37564272 Free PMC article.
References
-
- Gepts P. A comparison between crop domestication, classical plant breeding, and genetic engineering. Crop Sci. 2002;42:1780. doi: 10.2135/cropsci2002.1780. - DOI
-
- Peralta IE, Spooner DM, Knapp S, Anderson C. Taxonomy of wild tomatoes and their relatives (Solanum sect. Lycopersicoides, sect. Juglandifolia, sect. Lycopersicon; Solanaceae) Syst Bot Monogr. 2008;84:1–186.
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Other Literature Sources

