A human immune data-informed vaccine concept elicits strong and broad T-cell specificities associated with HIV-1 control in mice and macaques
- PMID: 25879820
- PMCID: PMC4336696
- DOI: 10.1186/s12967-015-0392-5
A human immune data-informed vaccine concept elicits strong and broad T-cell specificities associated with HIV-1 control in mice and macaques
Abstract
Background: None of the HIV T-cell vaccine candidates that have reached advanced clinical testing have been able to induce protective T cell immunity. A major reason for these failures may have been suboptimal T cell immunogen designs.
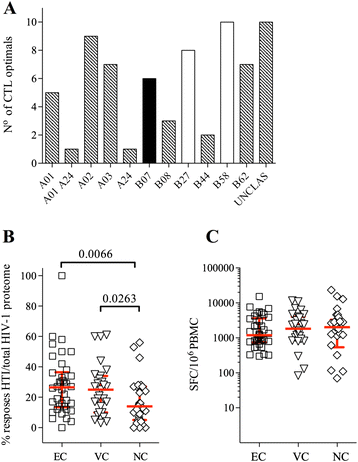
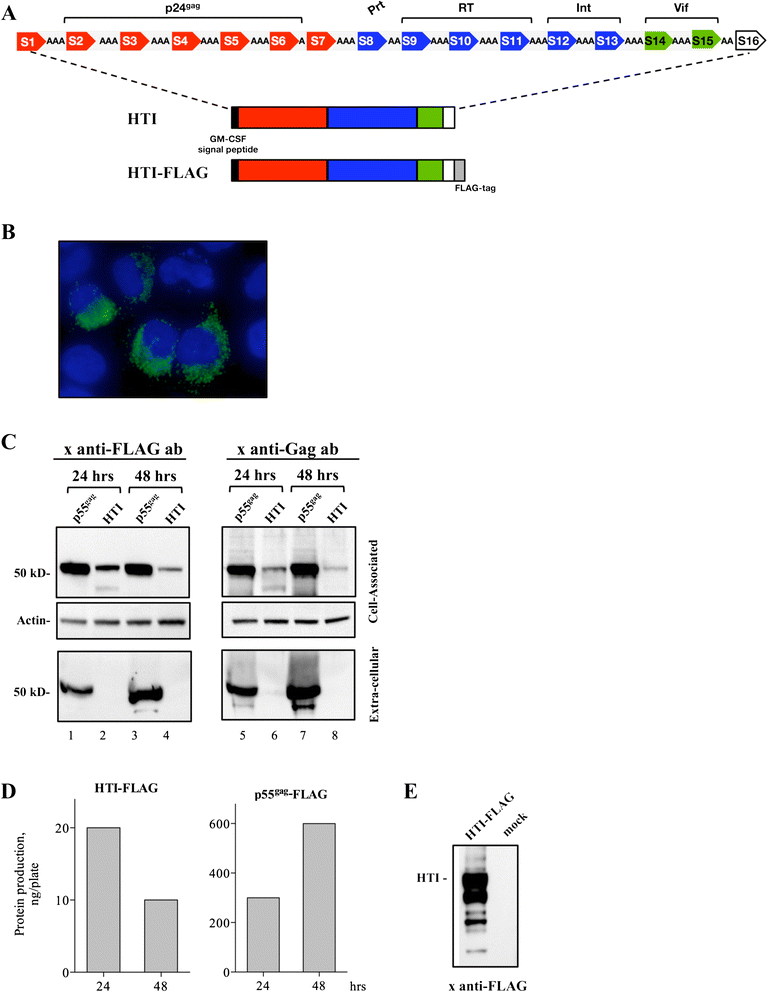
Methods: To overcome this problem, we used a novel immunogen design approach that is based on functional T cell response data from more than 1,000 HIV-1 clade B and C infected individuals and which aims to direct the T cell response to the most vulnerable sites of HIV-1.
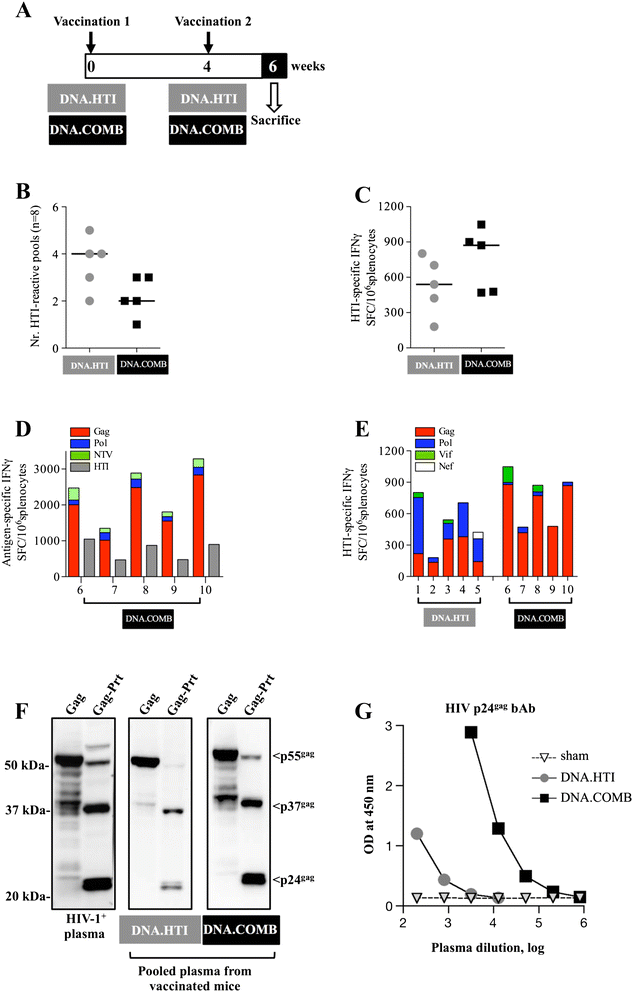
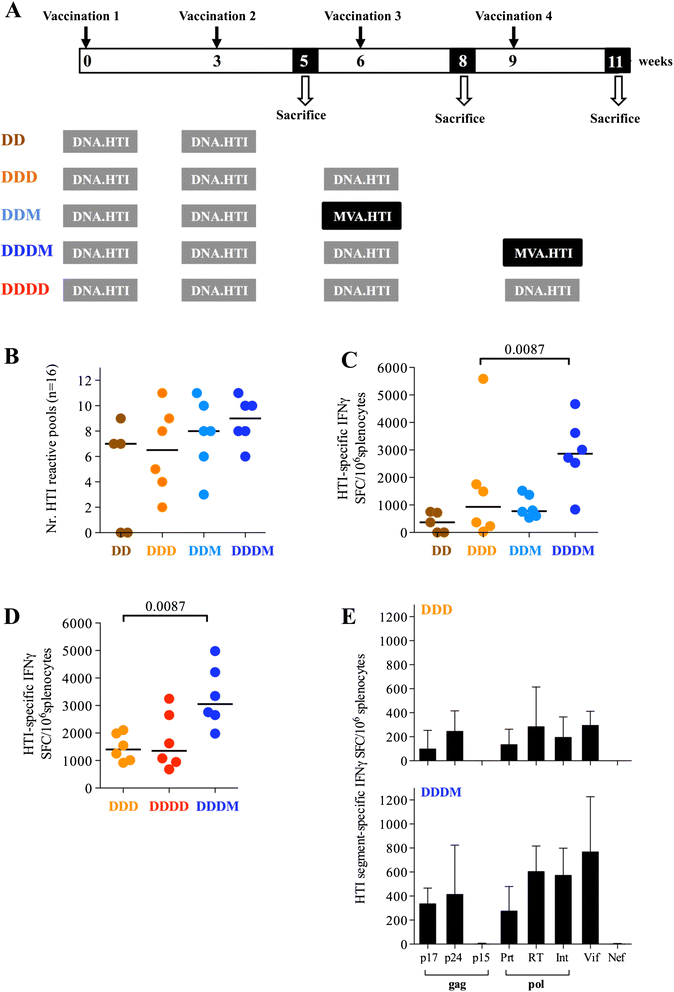
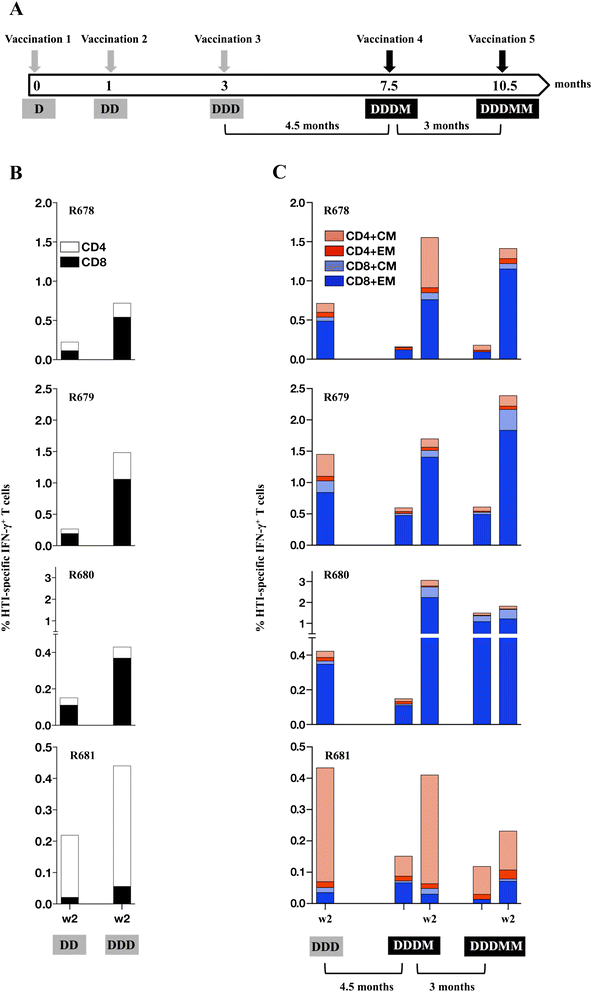
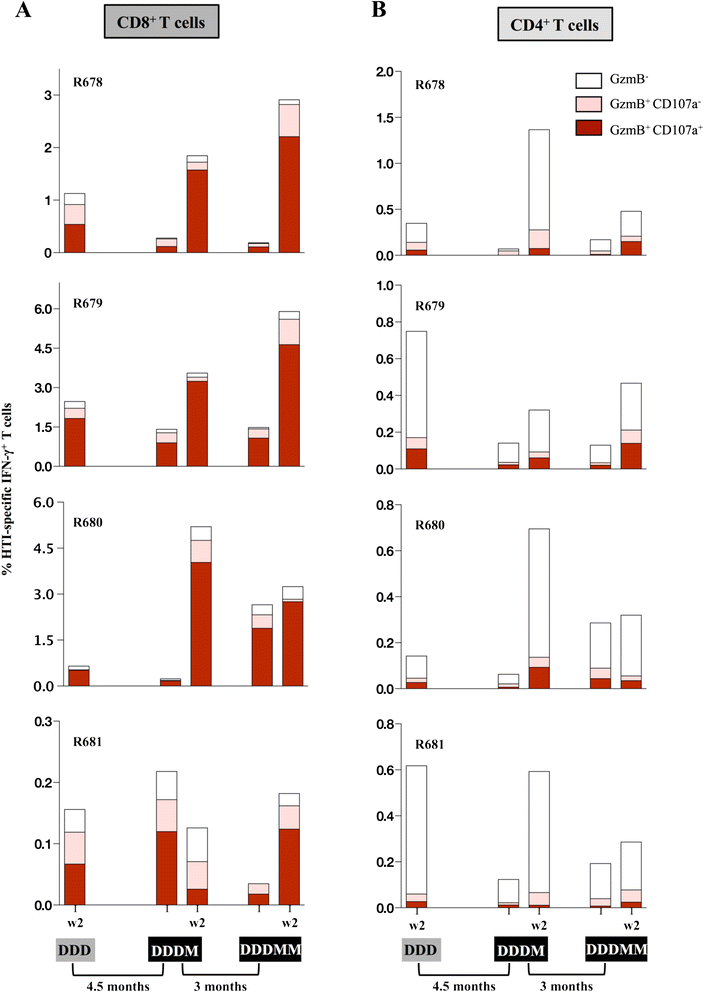
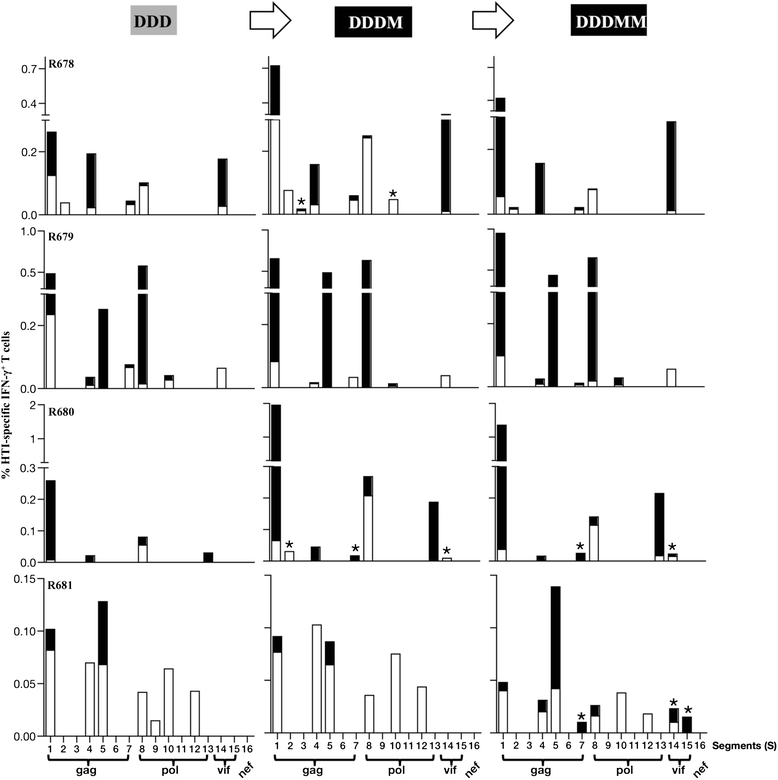
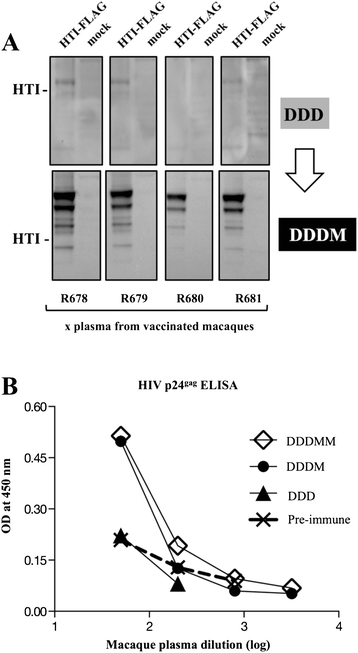
Results: Our approach identified 16 regions in Gag, Pol, Vif and Nef that were relatively conserved and predominantly targeted by individuals with reduced viral loads. These regions formed the basis of the HIVACAT T-cell Immunogen (HTI) sequence which is 529 amino acids in length, includes more than 50 optimally defined CD4(+) and CD8(+) T-cell epitopes restricted by a wide range of HLA class I and II molecules and covers viral sites where mutations led to a dramatic reduction in viral replicative fitness. In both, C57BL/6 mice and Indian rhesus macaques immunized with an HTI-expressing DNA plasmid (DNA.HTI) induced broad and balanced T-cell responses to several segments within Gag, Pol, and Vif. DNA.HTI induced robust CD4(+) and CD8(+) T cell responses that were increased by a booster vaccination using modified virus Ankara (MVA.HTI), expanding the DNA.HTI induced response to up to 3.2% IFN-γ T-cells in macaques. HTI-specific T cells showed a central and effector memory phenotype with a significant fraction of the IFN-γ(+) CD8(+) T cells being Granzyme B(+) and able to degranulate (CD107a(+)).
Conclusions: These data demonstrate the immunogenicity of a novel HIV-1 T cell vaccine concept that induced broadly balanced responses to vulnerable sites of HIV-1 while avoiding the induction of responses to potential decoy targets that may divert effective T-cell responses towards variable and less protective viral determinants.
Figures
Similar articles
-
Broad and cross-clade CD4+ T-cell responses elicited by a DNA vaccine encoding highly conserved and promiscuous HIV-1 M-group consensus peptides.PLoS One. 2012;7(9):e45267. doi: 10.1371/journal.pone.0045267. Epub 2012 Sep 18. PLoS One. 2012. PMID: 23028895 Free PMC article.
-
A Novel Immunogen Selectively Eliciting CD8+ T Cells but Not CD4+ T Cells Targeting Immunodeficiency Virus Antigens.J Virol. 2020 Mar 31;94(8):e01876-19. doi: 10.1128/JVI.01876-19. Print 2020 Mar 31. J Virol. 2020. PMID: 32024773 Free PMC article.
-
HIV/AIDS Vaccine Candidates Based on Replication-Competent Recombinant Poxvirus NYVAC-C-KC Expressing Trimeric gp140 and Gag-Derived Virus-Like Particles or Lacking the Viral Molecule B19 That Inhibits Type I Interferon Activate Relevant HIV-1-Specific B and T Cell Immune Functions in Nonhuman Primates.J Virol. 2017 Apr 13;91(9):e02182-16. doi: 10.1128/JVI.02182-16. Print 2017 May 1. J Virol. 2017. PMID: 28179536 Free PMC article.
-
Development of a DNA-MVA/HIVA vaccine for Kenya.Vaccine. 2002 May 6;20(15):1995-8. doi: 10.1016/s0264-410x(02)00085-3. Vaccine. 2002. PMID: 11983261 Review.
-
Identification of highly conserved and broadly cross-reactive HIV type 1 cytotoxic T lymphocyte epitopes as candidate immunogens for inclusion in Mycobacterium bovis BCG-vectored HIV vaccines.AIDS Res Hum Retroviruses. 2000 Sep 20;16(14):1433-43. doi: 10.1089/08892220050140982. AIDS Res Hum Retroviruses. 2000. PMID: 11018863 Review.
Cited by
-
The HIV-1 latent reservoir is largely sensitive to circulating T cells.Elife. 2020 Oct 6;9:e57246. doi: 10.7554/eLife.57246. Elife. 2020. PMID: 33021198 Free PMC article.
-
Phase I clinical trial of an intranodally administered mRNA-based therapeutic vaccine against HIV-1 infection.AIDS. 2018 Nov 13;32(17):2533-2545. doi: 10.1097/QAD.0000000000002026. AIDS. 2018. PMID: 30289805 Free PMC article. Clinical Trial.
-
Control of Heterologous Simian Immunodeficiency Virus SIVsmE660 Infection by DNA and Protein Coimmunization Regimens Combined with Different Toll-Like-Receptor-4-Based Adjuvants in Macaques.J Virol. 2018 Jul 17;92(15):e00281-18. doi: 10.1128/JVI.00281-18. Print 2018 Aug 1. J Virol. 2018. PMID: 29793957 Free PMC article.
-
Evaluation of the Thermal Stability of a Vaccine Prototype Based on Virus-like Particle Formulated HIV-1 Envelope.Vaccines (Basel). 2022 Mar 22;10(4):484. doi: 10.3390/vaccines10040484. Vaccines (Basel). 2022. PMID: 35455233 Free PMC article.
-
Induction of Broad and Polyfunctional HIV-1-Specific T Cell Responses by the Multiepitopic Protein TMEP-B Vectored by MVA Virus.Vaccines (Basel). 2019 Jun 29;7(3):57. doi: 10.3390/vaccines7030057. Vaccines (Basel). 2019. PMID: 31261918 Free PMC article.
References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Research Materials

