Structural Basis for the Inverted Repeat Preferences of mariner Transposases
- PMID: 25869132
- PMCID: PMC4505599
- DOI: 10.1074/jbc.M115.636704
Structural Basis for the Inverted Repeat Preferences of mariner Transposases
Abstract
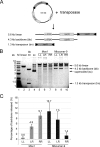
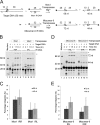
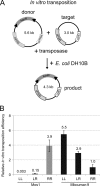
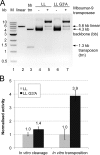
The inverted repeat (IR) sequences delimiting the left and right ends of many naturally active mariner DNA transposons are non-identical and have different affinities for their transposase. We have compared the preferences of two active mariner transposases, Mos1 and Mboumar-9, for their imperfect transposon IRs in each step of transposition: DNA binding, DNA cleavage, and DNA strand transfer. A 3.1 Å resolution crystal structure of the Mos1 paired-end complex containing the pre-cleaved left IR sequences reveals the molecular basis for the reduced affinity of the Mos1 transposase DNA-binding domain for the left IR as compared with the right IR. For both Mos1 and Mboumar-9, in vitro DNA transposition is most efficient when the preferred IR sequence is present at both transposon ends. We find that this is due to the higher efficiency of cleavage and strand transfer of the preferred transposon end. We show that the efficiency of Mboumar-9 transposition is improved almost 4-fold by changing the 3' base of the preferred Mboumar-9 IR from guanine to adenine. This preference for adenine at the reactive 3' end for both Mos1 and Mboumar-9 may be a general feature of mariner transposition.
Keywords: DNA recombination; DNA transposition; DNA-protein interaction; X-ray crystallography; molecular genetics; nucleic acid enzymology; phosphoryl transfer; structural biology.
© 2015 by The American Society for Biochemistry and Molecular Biology, Inc.
Figures


Similar articles
-
Regulation of mariner transposition: the peculiar case of Mos1.PLoS One. 2012;7(8):e43365. doi: 10.1371/journal.pone.0043365. Epub 2012 Aug 14. PLoS One. 2012. PMID: 22905263 Free PMC article.
-
Biochemical characterization and comparison of two closely related active mariner transposases.Biochemistry. 2014 Feb 4;53(4):682-9. doi: 10.1021/bi401193w. Epub 2014 Jan 21. Biochemistry. 2014. PMID: 24404958 Free PMC article.
-
Transposition of Mboumar-9: identification of a new naturally active mariner-family transposon.J Mol Biol. 2008 Oct 10;382(3):567-72. doi: 10.1016/j.jmb.2008.07.044. Epub 2008 Jul 23. J Mol Biol. 2008. PMID: 18675277 Free PMC article.
-
Assembly of the Tc1 and mariner transposition initiation complexes depends on the origins of their transposase DNA binding domains.Genetica. 2007 Jun;130(2):105-20. doi: 10.1007/s10709-006-0025-2. Epub 2006 Aug 16. Genetica. 2007. PMID: 16912840 Review.
-
Tn5 as a model for understanding DNA transposition.Mol Microbiol. 2003 Mar;47(5):1199-206. doi: 10.1046/j.1365-2958.2003.03382.x. Mol Microbiol. 2003. PMID: 12603728 Review.
Cited by
-
Distribution of Merlin in eukaryotes and first report of DNA transposons in kinetoplastid protists.PLoS One. 2021 May 6;16(5):e0251133. doi: 10.1371/journal.pone.0251133. eCollection 2021. PLoS One. 2021. PMID: 33956864 Free PMC article.
-
Targeted DNA transposition in vitro using a dCas9-transposase fusion protein.Nucleic Acids Res. 2019 Sep 5;47(15):8126-8135. doi: 10.1093/nar/gkz552. Nucleic Acids Res. 2019. PMID: 31429873 Free PMC article.
-
Affinities of Terminal Inverted Repeats to DNA Binding Domain of Transposase Affect the Transposition Activity of Bamboo Ppmar2 Mariner-Like Element.Int J Mol Sci. 2019 Jul 28;20(15):3692. doi: 10.3390/ijms20153692. Int J Mol Sci. 2019. PMID: 31357686 Free PMC article.
-
DNA Transposition at Work.Chem Rev. 2016 Oct 26;116(20):12758-12784. doi: 10.1021/acs.chemrev.6b00003. Epub 2016 May 17. Chem Rev. 2016. PMID: 27187082 Free PMC article. Review.
-
IS21 family transposase cleaved donor complex traps two right-handed superhelical crossings.Nat Commun. 2023 Apr 22;14(1):2335. doi: 10.1038/s41467-023-38071-x. Nat Commun. 2023. PMID: 37087515 Free PMC article.
References
-
- Delaurière L., Chénais B., Hardivillier Y., Gauvry L., Casse N. (2009) Mariner transposons as genetic tools in vertebrate cells. Genetica 137, 9–17 - PubMed
-
- Ammar I., Izsvák Z., Ivics Z. (2012) The Sleeping Beauty transposon toolbox. Methods Mol. Biol. 859, 229–240 - PubMed
-
- Ivics Z., Hackett P. B., Plasterk R. H., Izsvák Z. (1997) Molecular reconstruction of Sleeping Beauty, a Tc1-like transposon from fish, and its transposition in human cells. Cell 91, 501–510 - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

