GS-align for glycan structure alignment and similarity measurement
- PMID: 25857669
- PMCID: PMC4528633
- DOI: 10.1093/bioinformatics/btv202
GS-align for glycan structure alignment and similarity measurement
Abstract
Motivation: Glycans play critical roles in many biological processes, and their structural diversity is key for specific protein-glycan recognition. Comparative structural studies of biological molecules provide useful insight into their biological relationships. However, most computational tools are designed for protein structure, and despite their importance, there is no currently available tool for comparing glycan structures in a sequence order- and size-independent manner.
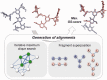
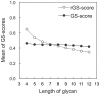
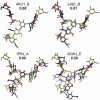
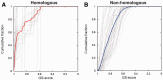
Results: A novel method, GS-align, is developed for glycan structure alignment and similarity measurement. GS-align generates possible alignments between two glycan structures through iterative maximum clique search and fragment superposition. The optimal alignment is then determined by the maximum structural similarity score, GS-score, which is size-independent. Benchmark tests against the Protein Data Bank (PDB) N-linked glycan library and PDB homologous/non-homologous N-glycoprotein sets indicate that GS-align is a robust computational tool to align glycan structures and quantify their structural similarity. GS-align is also applied to template-based glycan structure prediction and monosaccharide substitution matrix generation to illustrate its utility.
Availability and implementation: http://www.glycanstructure.org/gsalign.
Contact: wonpil@ku.edu
Supplementary information: Supplementary data are available at Bioinformatics online.
© The Author 2015. Published by Oxford University Press. All rights reserved. For Permissions, please e-mail: journals.permissions@oup.com.
Figures


Similar articles
-
Glycan Reader is improved to recognize most sugar types and chemical modifications in the Protein Data Bank.Bioinformatics. 2017 Oct 1;33(19):3051-3057. doi: 10.1093/bioinformatics/btx358. Bioinformatics. 2017. PMID: 28582506 Free PMC article.
-
LS-align: an atom-level, flexible ligand structural alignment algorithm for high-throughput virtual screening.Bioinformatics. 2018 Jul 1;34(13):2209-2218. doi: 10.1093/bioinformatics/bty081. Bioinformatics. 2018. PMID: 29462237 Free PMC article.
-
Restricted N-glycan conformational space in the PDB and its implication in glycan structure modeling.PLoS Comput Biol. 2013;9(3):e1002946. doi: 10.1371/journal.pcbi.1002946. Epub 2013 Mar 14. PLoS Comput Biol. 2013. PMID: 23516343 Free PMC article.
-
Computational glycoscience: characterizing the spatial and temporal properties of glycans and glycan-protein complexes.Curr Opin Struct Biol. 2010 Oct;20(5):575-83. doi: 10.1016/j.sbi.2010.07.005. Epub 2010 Aug 12. Curr Opin Struct Biol. 2010. PMID: 20708922 Free PMC article. Review.
-
Systems glycobiology: biochemical reaction networks regulating glycan structure and function.Glycobiology. 2011 Dec;21(12):1541-53. doi: 10.1093/glycob/cwr036. Epub 2011 Mar 24. Glycobiology. 2011. PMID: 21436236 Free PMC article. Review.
Cited by
-
Roles of glycans in interactions between gp120 and HIV broadly neutralizing antibodies.Glycobiology. 2016 Mar;26(3):251-60. doi: 10.1093/glycob/cwv101. Epub 2015 Nov 3. Glycobiology. 2016. PMID: 26537503 Free PMC article.
-
Glycan Reader is improved to recognize most sugar types and chemical modifications in the Protein Data Bank.Bioinformatics. 2017 Oct 1;33(19):3051-3057. doi: 10.1093/bioinformatics/btx358. Bioinformatics. 2017. PMID: 28582506 Free PMC article.
-
Three-Dimensional Structures of Carbohydrates and Where to Find Them.Int J Mol Sci. 2020 Oct 18;21(20):7702. doi: 10.3390/ijms21207702. Int J Mol Sci. 2020. PMID: 33081008 Free PMC article. Review.
-
CHARMM-GUI Glycan Modeler for modeling and simulation of carbohydrates and glycoconjugates.Glycobiology. 2019 Apr 1;29(4):320-331. doi: 10.1093/glycob/cwz003. Glycobiology. 2019. PMID: 30689864 Free PMC article.
-
Glycans in Virus-Host Interactions: A Structural Perspective.Front Mol Biosci. 2021 Jun 7;8:666756. doi: 10.3389/fmolb.2021.666756. eCollection 2021. Front Mol Biosci. 2021. PMID: 34164431 Free PMC article. Review.
References
-
- Aoki K.F., et al. . (2005) A score matrix to reveal the hidden links in glycans. Bioinformatics, 21, 1457–1463. - PubMed
-
- Aoki K.F., et al. . (2003) Efficient tree-matching methods for accurate carbohydrate database queries. Genome Inform., 14, 134–143. - PubMed
-
- Apweiler R., et al. . (1999) On the frequency of protein glycosylation, as deduced from analysis of the SWISS-PROT database. Biochim. Biophys. Acta, 1473, 4–8. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

