Human ACAP2 is a homolog of C. elegans CNT-1 that promotes apoptosis in cancer cells
- PMID: 25853217
- PMCID: PMC4614986
- DOI: 10.1080/15384101.2015.1026518
Human ACAP2 is a homolog of C. elegans CNT-1 that promotes apoptosis in cancer cells
Abstract
Activation of caspases is an integral part of the apoptotic cell death program. Collectively, these proteases target hundreds of substrates, leading to the hypothesis that apoptosis is "death by a thousand cuts". Recent work, however, has demonstrated that caspase cleavage of only a subset of these substrates directs apoptosis in the cell. One such example is C. elegans CNT-1, which is cleaved by CED-3 to generate a truncated form, tCNT-1, that acquires a potent phosphoinositide-binding activity and translocates to the plasma membrane where it inactivates AKT survival signaling. We report here that ACAP2, a homolog of C. elegans CNT-1, has a pro-apoptotic function and an identical phosphoinositide-binding pattern to that of tCNT-1, despite not being an apparent target of caspase cleavage. We show that knockdown of ACAP2 blocks apoptosis in cancer cells in response to the chemotherapeutic antimetabolite 5-fluorouracil and that ACAP2 expression is down-regulated in some esophageal cancers, leukemias and lymphomas. These results suggest that ACAP2 is a functional homolog of C. elegans CNT-1 and its inactivation or downregulation in human cells may contribute to cancer development.
Figures


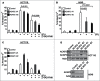
Similar articles
-
Caspase-activated phosphoinositide binding by CNT-1 promotes apoptosis by inhibiting the AKT pathway.Nat Struct Mol Biol. 2014 Dec;21(12):1082-90. doi: 10.1038/nsmb.2915. Epub 2014 Nov 10. Nat Struct Mol Biol. 2014. PMID: 25383666 Free PMC article.
-
Caenorhabditis elegans caspase homolog CSP-2 inhibits CED-3 autoactivation and apoptosis in germ cells.Cell Death Differ. 2009 Oct;16(10):1385-94. doi: 10.1038/cdd.2009.88. Epub 2009 Jul 3. Cell Death Differ. 2009. PMID: 19575016 Free PMC article.
-
Regulation of apoptosis by a Caenorhabditis elegans BNIP3 homolog.Oncogene. 1998 Nov 12;17(19):2525-30. doi: 10.1038/sj.onc.1202467. Oncogene. 1998. PMID: 9824163
-
2:1 Stoichiometry of the CED-4-CED-9 complex and the tetrameric CED-4: insights into the regulation of CED-3 activation.Cell Cycle. 2006 Jan;5(1):31-4. doi: 10.4161/cc.5.1.2263. Epub 2006 Jan 18. Cell Cycle. 2006. PMID: 16294007 Review.
-
Programmed cell death in Caenorhabditis elegans.Curr Opin Genet Dev. 1994 Aug;4(4):581-6. doi: 10.1016/0959-437x(94)90076-f. Curr Opin Genet Dev. 1994. PMID: 7950327 Review.
Cited by
-
Forensic age estimation from human blood using age-related microRNAs and circular RNAs markers.Front Genet. 2022 Nov 22;13:1031806. doi: 10.3389/fgene.2022.1031806. eCollection 2022. Front Genet. 2022. PMID: 36506317 Free PMC article.
-
Cancer-associated fibroblasts-derived exosomal miR-3656 promotes the development and progression of esophageal squamous cell carcinoma via the ACAP2/PI3K-AKT signaling pathway.Int J Biol Sci. 2021 Aug 27;17(14):3689-3701. doi: 10.7150/ijbs.62571. eCollection 2021. Int J Biol Sci. 2021. PMID: 34671193 Free PMC article.
-
Bioinformatics Analysis: The Regulatory Network of hsa_circ_0007843 and hsa_circ_0007331 in Colon Cancer.Biomed Res Int. 2021 Jul 23;2021:6662897. doi: 10.1155/2021/6662897. eCollection 2021. Biomed Res Int. 2021. PMID: 34337040 Free PMC article.
-
Integrative Analysis of Bulk RNA-Seq and Single-Cell RNA-Seq Unveils the Characteristics of the Immune Microenvironment and Prognosis Signature in Prostate Cancer.J Oncol. 2022 Jul 19;2022:6768139. doi: 10.1155/2022/6768139. eCollection 2022. J Oncol. 2022. PMID: 35909899 Free PMC article.
-
A novel lncRNA-mRNA-miRNA signature predicts recurrence and disease-free survival in cervical cancer.Braz J Med Biol Res. 2021 Sep 20;54(11):e11592. doi: 10.1590/1414-431X2021e11592. eCollection 2021. Braz J Med Biol Res. 2021. PMID: 34550275 Free PMC article.
References
-
- Shalini S, Dorstyn L, Dawar S, Kumar S. Old, new and emerging functions of caspases. Cell Death Differ [Internet] 2014. [cited 2014December21]; Available from: http://dx.doi.org/10.1038/cdd.2014.216; PMID:25526085 - DOI - PMC - PubMed
-
- Crawford ED, Wells JA. Caspase substrates and cellular remodeling. Annu Rev Biochem [Internet] 2011. [cited 2014October29]; 80:1055-87. Available from: http://www.annualreviews.org/doi/abs/10.1146/annurev-biochem-061809-121639; PMID:21456965; http://dx.doi.org/10.1146/annurev-biochem-061809-121639 - DOI - DOI - PubMed
-
- Poreba M, Strózyk A, Salvesen GS, Drag M. Caspase substrates and inhibitors. Cold Spring Harb Perspect Biol [Internet] 2013. [cited 2014November28]; 5:a008680. Available from: http://cshperspectives.cshlp.org/content/5/8/a008680.long; PMID:23788633; http://dx.doi.org/10.1101/cshperspect.a008680 - DOI - PMC - PubMed
-
- Stroh C, Schulze-Osthoff K. Death by a thousand cuts: an ever increasing list of caspase substrates. Cell Death Differ [Internet] 1998. [cited 2015January9]; 5:997-1000. Available from: http://www.ncbi.nlm.nih.gov/pubmed/9894605; PMID:9894605; http://dx.doi.org/10.1038/sj.cdd.4400451 - DOI - PubMed
-
- Crawford ED, Seaman JE, Barber AE, David DC, Babbitt PC, Burlingame AL, Wells JA. Conservation of caspase substrates across metazoans suggests hierarchical importance of signaling pathways over specific targets and cleavage site motifs in apoptosis. Cell Death Differ [Internet] 2012. [cited 2015January9]; 19:2040-8. Available from: http://www.pubmedcentral.nih.gov/articlerender.fcgi?artid=3504717&tool=p...; PMID:22918439; http://dx.doi.org/10.1038/cdd.2012.99 - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
