Genomic analyses reveal mutational signatures and frequently altered genes in esophageal squamous cell carcinoma
- PMID: 25839328
- PMCID: PMC4385186
- DOI: 10.1016/j.ajhg.2015.02.017
Genomic analyses reveal mutational signatures and frequently altered genes in esophageal squamous cell carcinoma
Erratum in
- Am J Hum Genet. 2015 Nov 5;97(5):769
-
Genomic Analyses Reveal Mutational Signatures and Frequently Altered Genes in Esophageal Squamous Cell Carcinoma.Am J Hum Genet. 2020 Aug 6;107(2):375. doi: 10.1016/j.ajhg.2020.07.008. Am J Hum Genet. 2020. PMID: 32763191 Free PMC article. No abstract available.
-
Genomic Analyses Reveal Mutational Signatures and Frequently Altered Genes in Esophageal Squamous Cell Carcinoma.Am J Hum Genet. 2020 Sep 3;107(3):579. doi: 10.1016/j.ajhg.2020.08.012. Am J Hum Genet. 2020. PMID: 32888509 Free PMC article. No abstract available.
Abstract
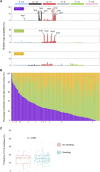
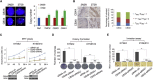
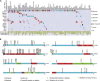
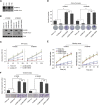
Esophageal squamous cell carcinoma (ESCC) is one of the most common cancers worldwide and the fourth most lethal cancer in China. However, although genomic studies have identified some mutations associated with ESCC, we know little of the mutational processes responsible. To identify genome-wide mutational signatures, we performed either whole-genome sequencing (WGS) or whole-exome sequencing (WES) on 104 ESCC individuals and combined our data with those of 88 previously reported samples. An APOBEC-mediated mutational signature in 47% of 192 tumors suggests that APOBEC-catalyzed deamination provides a source of DNA damage in ESCC. Moreover, PIK3CA hotspot mutations (c.1624G>A [p.Glu542Lys] and c.1633G>A [p.Glu545Lys]) were enriched in APOBEC-signature tumors, and no smoking-associated signature was observed in ESCC. In the samples analyzed by WGS, we identified focal (<100 kb) amplifications of CBX4 and CBX8. In our combined cohort, we identified frequent inactivating mutations in AJUBA, ZNF750, and PTCH1 and the chromatin-remodeling genes CREBBP and BAP1, in addition to known mutations. Functional analyses suggest roles for several genes (CBX4, CBX8, AJUBA, and ZNF750) in ESCC. Notably, high activity of hedgehog signaling and the PI3K pathway in approximately 60% of 104 ESCC tumors indicates that therapies targeting these pathways might be particularly promising strategies for ESCC. Collectively, our data provide comprehensive insights into the mutational signatures of ESCC and identify markers for early diagnosis and potential therapeutic targets.
Copyright © 2015 The Authors. Published by Elsevier Inc. All rights reserved.
Figures

Similar articles
-
Genomic analyses reveal FAM84B and the NOTCH pathway are associated with the progression of esophageal squamous cell carcinoma.Gigascience. 2016 Jan 11;5:1. doi: 10.1186/s13742-015-0107-0. eCollection 2016. Gigascience. 2016. PMID: 26759717 Free PMC article.
-
Genomic Landscape of Esophageal Squamous Cell Carcinoma in a Japanese Population.Gastroenterology. 2016 May;150(5):1171-1182. doi: 10.1053/j.gastro.2016.01.035. Epub 2016 Feb 10. Gastroenterology. 2016. PMID: 26873401
-
Germline and somatic variations influence the somatic mutational signatures of esophageal squamous cell carcinomas in a Chinese population.BMC Genomics. 2018 Jul 16;19(1):538. doi: 10.1186/s12864-018-4906-4. BMC Genomics. 2018. PMID: 30012096 Free PMC article.
-
Genomic characterization of esophageal squamous cell carcinoma: Insights from next-generation sequencing.World J Gastroenterol. 2016 Feb 21;22(7):2284-93. doi: 10.3748/wjg.v22.i7.2284. World J Gastroenterol. 2016. PMID: 26900290 Free PMC article. Review.
-
Cancers of the upper gastro-intestinal tract: a review of somatic mutation distributions.Arch Iran Med. 2014 Apr;17(4):286-92. Arch Iran Med. 2014. PMID: 24724606 Review.
Cited by
-
Hedgehog signaling mechanism and role in cancer.Semin Cancer Biol. 2022 Oct;85:107-122. doi: 10.1016/j.semcancer.2021.04.003. Epub 2021 Apr 6. Semin Cancer Biol. 2022. PMID: 33836254 Free PMC article. Review.
-
Global research trend of esophageal squamous cell carcinoma from 2012 to 2022: a bibliometric analysis.Front Oncol. 2022 Aug 11;12:977935. doi: 10.3389/fonc.2022.977935. eCollection 2022. Front Oncol. 2022. PMID: 36033465 Free PMC article.
-
Human umbilical cord mesenchymal stem cells-derived exosomes deliver microRNA-375 to downregulate ENAH and thus retard esophageal squamous cell carcinoma progression.J Exp Clin Cancer Res. 2020 Jul 22;39(1):140. doi: 10.1186/s13046-020-01631-w. J Exp Clin Cancer Res. 2020. PMID: 32698859 Free PMC article.
-
Identification of HLA-A2-Restricted Mutant Epitopes from Neoantigens of Esophageal Squamous Cell Carcinoma.Vaccines (Basel). 2021 Oct 1;9(10):1118. doi: 10.3390/vaccines9101118. Vaccines (Basel). 2021. PMID: 34696226 Free PMC article.
-
Transcriptome profiling and analysis of patients with esophageal squamous cell carcinoma from Kazakhstan.Front Genet. 2024 Mar 18;15:1249751. doi: 10.3389/fgene.2024.1249751. eCollection 2024. Front Genet. 2024. PMID: 38562378 Free PMC article.
References
-
- Pennathur A., Gibson M.K., Jobe B.A., Luketich J.D. Oesophageal carcinoma. Lancet. 2013;381:400–412. - PubMed
-
- Engel L.S., Chow W.H., Vaughan T.L., Gammon M.D., Risch H.A., Stanford J.L., Schoenberg J.B., Mayne S.T., Dubrow R., Rotterdam H. Population attributable risks of esophageal and gastric cancers. J. Natl. Cancer Inst. 2003;95:1404–1413. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Miscellaneous

