Essential roles for the splicing regulator nSR100/SRRM4 during nervous system development
- PMID: 25838543
- PMCID: PMC4387716
- DOI: 10.1101/gad.256115.114
Essential roles for the splicing regulator nSR100/SRRM4 during nervous system development
Abstract
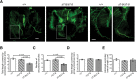
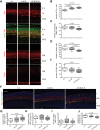
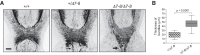
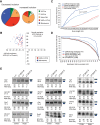
Alternative splicing (AS) generates vast transcriptomic complexity in the vertebrate nervous system. However, the extent to which trans-acting splicing regulators and their target AS regulatory networks contribute to nervous system development is not well understood. To address these questions, we generated mice lacking the vertebrate- and neural-specific Ser/Arg repeat-related protein of 100 kDa (nSR100/SRRM4). Loss of nSR100 impairs development of the central and peripheral nervous systems in part by disrupting neurite outgrowth, cortical layering in the forebrain, and axon guidance in the corpus callosum. Accompanying these developmental defects are widespread changes in AS that primarily result in shifts to nonneural patterns for different classes of splicing events. The main component of the altered AS program comprises 3- to 27-nucleotide (nt) neural microexons, an emerging class of highly conserved AS events associated with the regulation of protein interaction networks in developing neurons and neurological disorders. Remarkably, inclusion of a 6-nt, nSR100-activated microexon in Unc13b transcripts is sufficient to rescue a neuritogenesis defect in nSR100 mutant primary neurons. These results thus reveal critical in vivo neurodevelopmental functions of nSR100 and further link these functions to a conserved program of neuronal microexon splicing.
Keywords: SR proteins; alternative splicing; microexons; nervous system development.
© 2015 Quesnel-Vallières et al.; Published by Cold Spring Harbor Laboratory Press.
Figures

Similar articles
-
A highly conserved program of neuronal microexons is misregulated in autistic brains.Cell. 2014 Dec 18;159(7):1511-23. doi: 10.1016/j.cell.2014.11.035. Cell. 2014. PMID: 25525873 Free PMC article.
-
Cross-regulation between an alternative splicing activator and a transcription repressor controls neurogenesis.Mol Cell. 2011 Sep 2;43(5):843-50. doi: 10.1016/j.molcel.2011.08.014. Mol Cell. 2011. PMID: 21884984
-
Neuronal-specific microexon splicing of TAF1 mRNA is directly regulated by SRRM4/nSR100.RNA Biol. 2020 Jan;17(1):62-74. doi: 10.1080/15476286.2019.1667214. Epub 2019 Sep 27. RNA Biol. 2020. PMID: 31559909 Free PMC article.
-
Alternative Splicing in the Mammalian Nervous System: Recent Insights into Mechanisms and Functional Roles.Neuron. 2015 Jul 1;87(1):14-27. doi: 10.1016/j.neuron.2015.05.004. Neuron. 2015. PMID: 26139367 Review.
-
Microexons: at the nexus of nervous system development, behaviour and autism spectrum disorder.Curr Opin Genet Dev. 2020 Dec;65:22-33. doi: 10.1016/j.gde.2020.03.007. Epub 2020 Jun 11. Curr Opin Genet Dev. 2020. PMID: 32535349 Review.
Cited by
-
Circular RNA in cancer.Nat Rev Cancer. 2024 Sep;24(9):597-613. doi: 10.1038/s41568-024-00721-7. Epub 2024 Jul 29. Nat Rev Cancer. 2024. PMID: 39075222 Review.
-
Precise temporal regulation of alternative splicing during neural development.Nat Commun. 2018 Jun 6;9(1):2189. doi: 10.1038/s41467-018-04559-0. Nat Commun. 2018. PMID: 29875359 Free PMC article.
-
Specialization of the photoreceptor transcriptome by Srrm3-dependent microexons is required for outer segment maintenance and vision.Proc Natl Acad Sci U S A. 2022 Jul 19;119(29):e2117090119. doi: 10.1073/pnas.2117090119. Epub 2022 Jul 12. Proc Natl Acad Sci U S A. 2022. PMID: 35858306 Free PMC article.
-
Characterising the RNA-binding protein atlas of the mammalian brain uncovers RBM5 misregulation in mouse models of Huntington's disease.Nat Commun. 2023 Jul 19;14(1):4348. doi: 10.1038/s41467-023-39936-x. Nat Commun. 2023. PMID: 37468457 Free PMC article.
-
An RNA Switch of a Large Exon of Ninein Is Regulated by the Neural Stem Cell Specific-RNA Binding Protein, Qki5.Int J Mol Sci. 2019 Feb 26;20(5):1010. doi: 10.3390/ijms20051010. Int J Mol Sci. 2019. PMID: 30813567 Free PMC article.
References
-
- Akbarian S, Bunney WE Jr, Potkin SG, Wigal SB, Hagman JO, Sandman CA, Jones EG. 1993. Altered distribution of nicotinamide-adenine dinucleotide phosphate-diaphorase cells in frontal lobe of schizophrenics implies disturbances of cortical development. Arch Gen Psychiatry 50: 169–177. - PubMed
-
- Barbosa-Morais NL, Irimia M, Pan Q, Xiong HY, Gueroussov S, Lee LJ, Slobodeniuc V, Kutter C, Watt S, Colak R, et al.2012. The evolutionary landscape of alternative splicing in vertebrate species. Science 338: 1587–1593. - PubMed
-
- Boucard AA, Chubykin AA, Comoletti D, Taylor P, Sudhof TC. 2005. A splice code for trans-synaptic cell adhesion mediated by binding of neuroligin 1 to α- and β-neurexins. Neuron 48: 229–236. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials
