A universal mariner transposon system for forward genetic studies in the genus Clostridium
- PMID: 25836262
- PMCID: PMC4383383
- DOI: 10.1371/journal.pone.0122411
A universal mariner transposon system for forward genetic studies in the genus Clostridium
Abstract
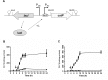
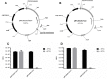
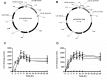
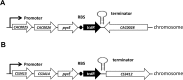
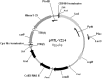
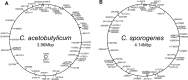
DNA transposons represent an essential tool in the armoury of the molecular microbiologist. We previously developed a catP-based mini transposon system for Clostridium difficile in which the expression of the transposase gene was dependent on a sigma factor unique to C. difficile, TcdR. Here we have shown that the host range of the transposon is easily extended through the rapid chromosomal insertion of the tcdR gene at the pyrE locus of the intended clostridial target using Allele-Coupled Exchange (ACE). To increase the effectiveness of the system, a novel replicon conditional for plasmid maintenance was developed, which no longer supports the effective retention of the transposon delivery vehicle in the presence of the inducer isopropyl β-D-1-thiogalactopyranoside (IPTG). As a consequence, those thiamphenicol resistant colonies that arise in clostridial recipients, following plating on agar medium supplemented with IPTG, are almost exclusively due to insertion of the mini transposon into the genome. The system has been exemplified in both Clostridium acetobutylicum and Clostridium sporogenes, where transposon insertion has been shown to be entirely random. Moreover, appropriate screening of both libraries resulted in the isolation of auxotrophic mutants as well as cells deficient in spore formation/germination. This strategy is capable of being implemented in any Clostridium species.
Conflict of interest statement
Figures
Similar articles
-
A roadmap for gene system development in Clostridium.Anaerobe. 2016 Oct;41:104-112. doi: 10.1016/j.anaerobe.2016.05.011. Epub 2016 May 24. Anaerobe. 2016. PMID: 27234263 Free PMC article. Review.
-
Development of an inducible transposon system for efficient random mutagenesis in Clostridium acetobutylicum.FEMS Microbiol Lett. 2016 Apr;363(8):fnw065. doi: 10.1093/femsle/fnw065. Epub 2016 Mar 20. FEMS Microbiol Lett. 2016. PMID: 27001972 Free PMC article.
-
A mariner-based transposon system for in vivo random mutagenesis of Clostridium difficile.Appl Environ Microbiol. 2010 Feb;76(4):1103-9. doi: 10.1128/AEM.02525-09. Epub 2009 Dec 18. Appl Environ Microbiol. 2010. PMID: 20023081 Free PMC article.
-
Transposon mutagenesis of Clostridium acetobutylicum P262: isolation and characterization of solvent deficient and metronidazole resistant mutants.FEMS Microbiol Lett. 1993 Dec 15;114(3):343-8. doi: 10.1111/j.1574-6968.1993.tb06596.x. FEMS Microbiol Lett. 1993. PMID: 8288111
-
The clostridial mobilisable transposons.Cell Mol Life Sci. 2002 Dec;59(12):2033-43. doi: 10.1007/s000180200003. Cell Mol Life Sci. 2002. PMID: 12568329 Free PMC article. Review.
Cited by
-
Gas Fermentation-A Flexible Platform for Commercial Scale Production of Low-Carbon-Fuels and Chemicals from Waste and Renewable Feedstocks.Front Microbiol. 2016 May 11;7:694. doi: 10.3389/fmicb.2016.00694. eCollection 2016. Front Microbiol. 2016. PMID: 27242719 Free PMC article. Review.
-
A roadmap for gene system development in Clostridium.Anaerobe. 2016 Oct;41:104-112. doi: 10.1016/j.anaerobe.2016.05.011. Epub 2016 May 24. Anaerobe. 2016. PMID: 27234263 Free PMC article. Review.
-
First Complete Genome Sequence of Clostridium sporogenes DSM 795T, a Nontoxigenic Surrogate for Clostridium botulinum, Determined Using PacBio Single-Molecule Real-Time Technology.Genome Announc. 2015 Jul 30;3(4):e00832-15. doi: 10.1128/genomeA.00832-15. Genome Announc. 2015. PMID: 26227598 Free PMC article.
-
Development of an inducible transposon system for efficient random mutagenesis in Clostridium acetobutylicum.FEMS Microbiol Lett. 2016 Apr;363(8):fnw065. doi: 10.1093/femsle/fnw065. Epub 2016 Mar 20. FEMS Microbiol Lett. 2016. PMID: 27001972 Free PMC article.
-
Required Gene Set for Autotrophic Growth of Clostridium autoethanogenum.Appl Environ Microbiol. 2022 Apr 12;88(7):e0247921. doi: 10.1128/aem.02479-21. Epub 2022 Mar 14. Appl Environ Microbiol. 2022. PMID: 35285680 Free PMC article.
References
-
- Brazier JS (2008) Clostridium difficile: from obscurity to superbug. Br J Biomed Sci 65: 39–44. - PubMed
-
- Dürre P (2005) Handbook on clostridia Boca Raton: Taylor & Francis. 903 p. p.
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials
Miscellaneous

